5SRS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z2614735107 - (R) and (S) isomers | | Descriptor: | 3-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SRV
 
 | |
5SRU
 
 | |
5SRT
 
 | |
5SSA
 
 | |
5SSQ
 
 | |
1I2R
 
 | |
1I2P
 
 | |
1I2Q
 
 | |
1I2O
 
 | |
1I2N
 
 | |
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1HF6
 
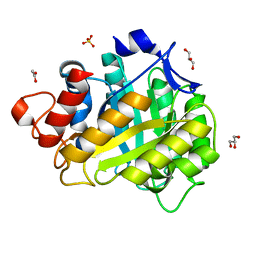 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
