3AW5
 
 | | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AXB
 
 | | Structure of a dye-linked L-proline dehydrogenase from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, PROLINE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K, Hara, Y. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
2ZPO
 
 | | Crystal Structure of Green Turtle Egg White Ribonuclease | | Descriptor: | GLYCEROL, Ribonuclease, SULFATE ION | | Authors: | Katekaew, S, Kakuta, Y, Torikata, T, Kimura, M, Yoneda, K, Araki, T. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Newly Found Green Turtle Egg White Ribonuclease
To be Published
|
|
3AW9
 
 | |
3AZ4
 
 | | Crystal structure of Co/O-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ5
 
 | | Crystal structure of Pt/O-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ6
 
 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ7
 
 | | Crystal structure of Pt/T-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION, SODIUM ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
8J1C
 
 | |
8J1G
 
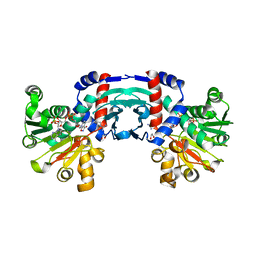 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
1VCV
 
 | |
1WZU
 
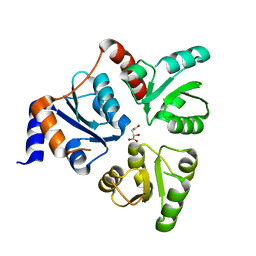 | | Crystal structure of quinolinate synthase (nadA) | | Descriptor: | D-MALATE, Quinolinate synthetase A | | Authors: | Sakuraba, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NAD Biosynthetic Enzyme Quinolinate Synthase
J.Biol.Chem., 280, 2005
|
|
4YSV
 
 | |
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WYA
 
 | | Structure of amino acid racemase, 2.65 A | | Descriptor: | (2S,3S)-3-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pentanoic acid, DIMETHYL SULFOXIDE, Isoleucine 2-epimerase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WYF
 
 | | Structure of amino acid racemase, 2.12 A | | Descriptor: | CADMIUM ION, Isoleucine 2-epimerase, N-[O-PHOSPHONO-PYRIDOXYL]-ISOLEUCINE | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7VNO
 
 | | Structure of aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 454aa long hypothetical 4-aminobutyrate aminotransferase, GLYCEROL, ... | | Authors: | Sakuraba, H, Ohshida, T, Ohshima, T. | | Deposit date: | 2021-10-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
7VO1
 
 | | Structure of aminotransferase-substrate complex | | Descriptor: | 454aa long hypothetical 4-aminobutyrate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Sakuraba, H, Ohshida, T, Ohshima, T. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
7VNT
 
 | | Structure of aminotransferase-substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 454aa long hypothetical 4-aminobutyrate aminotransferase, GLYCEROL, ... | | Authors: | Sakuraba, H, Ohshida, T, Ohshima, T. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
2ZVR
 
 | |
