3RA6
 
 | |
3VB6
 
 | | Crystal structure of SARS-CoV 3C-like protease with C6Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C6Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB3
 
 | | Crystal structure of SARS-CoV 3C-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, DI(HYDROXYETHYL)ETHER | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3RA5
 
 | | Crystal structure of T. celer L30e E6A/R92A variant | | Descriptor: | 50S ribosomal protein L30e, SULFATE ION | | Authors: | Chan, C.H, Wong, K.B. | | Deposit date: | 2011-03-27 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing salt-bridge enhances protein thermostability by reducing the heat capacity change of unfolding
Plos One, 6, 2011
|
|
7VB2
 
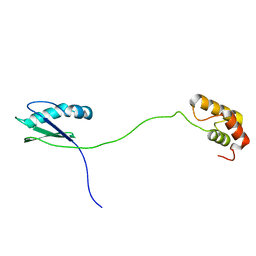 | | Solution structure of human ribosomal protein uL11 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Lee, K.M, Wong, K.B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The flexible N-terminal motif of uL11 unique to eukaryotic ribosomes interacts with P-complex and facilitates protein translation.
Nucleic Acids Res., 50, 2022
|
|
1GIU
 
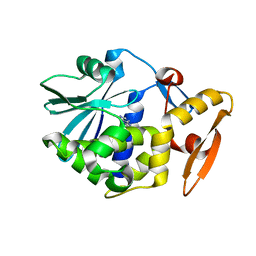 | | A TRICHOSANTHIN(TCS) MUTANT(E85R) COMPLEX STRUCTURE WITH ADENINE | | Descriptor: | ADENINE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
1GIS
 
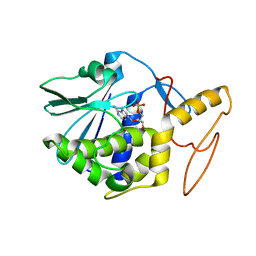 | | A TRICHOSANTHIN(TCS) MUTANT(E85Q) COMPLEX STRUCTURE WITH 2'-DEOXY-ADENOSIN-5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
3TNV
 
 | |
3TOQ
 
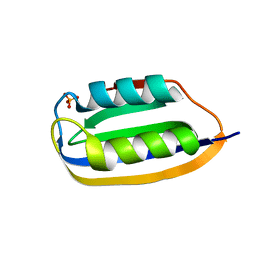 | |
