9ISE
 
 | | Human MTHFD1 in complex with LY374571 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-tetrahydrofolate synthase, cytoplasmic, ... | | Authors: | Lee, L.C, Wu, S.Y. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Potent and Selective Inhibitors of Methylenetetrahydrofolate Dehydrogenase 2 for Targeting Acute Myeloid Leukemia: SAR, Structural Insights, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
9IT3
 
 | | Human MTHFD2 in complex with compound 16e | | Descriptor: | (2~{S})-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1~{H}-pyrimidin-5-yl]carbamoylamino]-3-chloranyl-phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Lee, L.C, Wu, S.Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of Potent and Selective Inhibitors of Methylenetetrahydrofolate Dehydrogenase 2 for Targeting Acute Myeloid Leukemia: SAR, Structural Insights, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
9ITD
 
 | | Human MTHFD1 in complex with compound 16a | | Descriptor: | (2~{S})-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1~{H}-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, C-1-tetrahydrofolate synthase, cytoplasmic, ... | | Authors: | Lee, L.C, Wu, S.Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Development of Potent and Selective Inhibitors of Methylenetetrahydrofolate Dehydrogenase 2 for Targeting Acute Myeloid Leukemia: SAR, Structural Insights, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
9ISR
 
 | | Human MTHFD1 in complex with compound 16g | | Descriptor: | (2~{S})-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1~{H}-pyrimidin-5-yl]carbamoylamino]-3-chloranyl-phenyl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, C-1-tetrahydrofolate synthase, cytoplasmic, ... | | Authors: | Lee, L.C, Wu, S.Y. | | Deposit date: | 2024-07-18 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of Potent and Selective Inhibitors of Methylenetetrahydrofolate Dehydrogenase 2 for Targeting Acute Myeloid Leukemia: SAR, Structural Insights, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
1ES1
 
 | | CRYSTAL STRUCTURE OF VAL61HIS MUTANT OF TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
1EHB
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT TRYPSIN-SOLUBILIZED FRAGMENT OF CYTOCHROME B5 | | Descriptor: | PROTEIN (CYTOCHROME B5), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, J, Gan, J.-H, Xia, Z.-X, Wang, Y.-H, Wang, W.-H, Xue, L.-L, Xie, Y, Huang, Z.-X. | | Deposit date: | 2000-02-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b(5) and the structural comparison with Val61His mutant.
Proteins, 40, 2000
|
|
9JIU
 
 | | Ferritin mutant R63MeHis | | Descriptor: | Ferritin heavy chain, HEXAETHYLENE GLYCOL, SODIUM ION | | Authors: | Tsou, J.-C, Huang, K.-F, Ko, T.-P, Wang, Y.-S. | | Deposit date: | 2024-09-12 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Site-Specific Histidine Aza-Michael Addition in Proteins Enabled by a Ferritin-Based Metalloenzyme.
J.Am.Chem.Soc., 146, 2024
|
|
1NX7
 
 | |
1M20
 
 | | Crystal Structure of F35Y Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yao, P, Wu, J, Wang, Y.-H, Sun, B.-Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-06-20 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography, CD and kinetic studies revealed the essence of the abnormal behaviors of the cytochrome b5 Phe35-->Tyr mutant.
Eur.J.Biochem., 269, 2002
|
|
1SH4
 
 | |
1U9U
 
 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4YL9
 
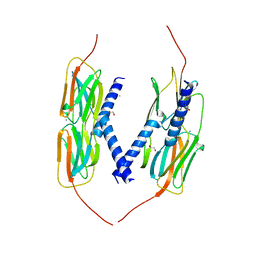 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
4YLC
 
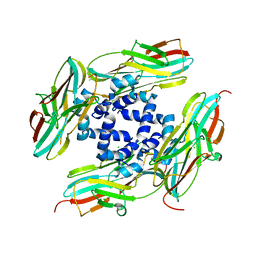 | |
4YLB
 
 | |
8KE2
 
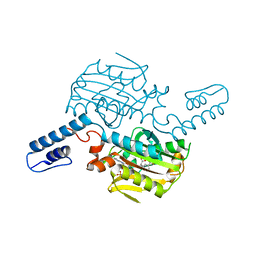 | | PylRS C-terminus domain mutant bound with L-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2S)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19639969 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE3
 
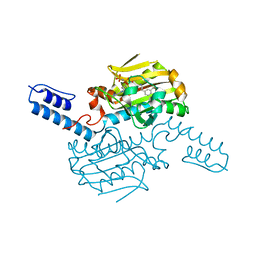 | | PylRS C-terminus domain mutant bound with D-3-trifluoromethylphenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-[3-(trifluoromethyl)phenyl]propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89980984 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE5
 
 | | PylRS C-terminus domain mutant bound with D-3-chlorophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-chlorophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.900073 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE6
 
 | | PylRS C-terminus domain mutant bound with L-3-chlorophenylalanine and AMPNP | | Descriptor: | 3-CHLORO-L-PHENYLALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89570856 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE1
 
 | | PylRS C-terminus domain mutant bound with L-3-bromophenylalanine and AMPNP | | Descriptor: | 3-bromo-L-phenylalanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50081539 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
8KE4
 
 | | PylRS C-terminus domain mutant bound with D-3-bromophenylalanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(3-bromophenyl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75050962 Å) | | Cite: | Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.
Front Genet, 14, 2023
|
|
6LY6
 
 | | PylRS C-terminus domain mutant bound with 3-(1-Naphthyl)-L-alanine and AMPNP | | Descriptor: | MAGNESIUM ION, NAPHTHALEN-2-YL-3-ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5001328 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LY3
 
 | | PylRS C-terminus domain mutant bound with 3-Benzothienyl-L-alanine and AMPNP | | Descriptor: | 3-(1-benzothiophen-3-yl)-L-alanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89590144 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LYA
 
 | | PylRS C-terminus domain mutant bound with 1-Methyl-L-tryptophan and AMPNP | | Descriptor: | 1,2-ETHANEDIOL, 1-Methyl-L-tryptophan, MAGNESIUM ION, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.590702 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LYB
 
 | | PylRS C-terminus domain mutant in complex with 3-Benzothienyl-D-alanine and AMPNP | | Descriptor: | (2R)-2-azanyl-3-(1-benzothiophen-3-yl)propanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.90366471 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
