3ENU
 
 | |
4NBI
 
 | | D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 1.86 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-(D-tyrosylamino)adenosine, D-tyrosyl-tRNA(Tyr) deacylase, TRIETHYLENE GLYCOL | | Authors: | Ahmad, S, Routh, S.B, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanism of chiral proofreading during translation of the genetic code.
Elife, 2, 2013
|
|
2HL0
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HL2
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HKZ
 
 | |
2HL1
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
5H4E
 
 | |
5HT9
 
 | |
5ZRN
 
 | |
5J61
 
 | |
3T5C
 
 | |
3T5A
 
 | |
3SNY
 
 | |
3SO1
 
 | |
3T5B
 
 | |
3PD4
 
 | |
3PD3
 
 | |
3PD2
 
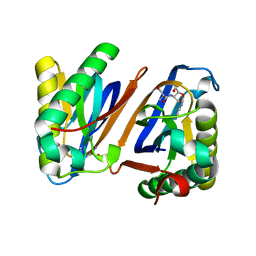 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kamarthapu, V, Kruparani, S.P, Sankaranarayanan, R. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanistic insights into cognate substrate discrimination during proofreading in translation
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PD5
 
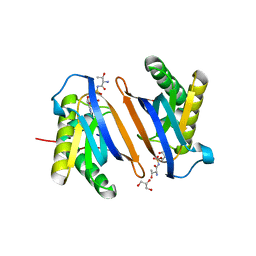 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of threonyl-adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kamarthapu, V, Kruparani, S.P, Sankaranarayanan, R. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mechanistic insights into cognate substrate discrimination during proofreading in translation
Proc.Natl.Acad.Sci.USA, 2010
|
|
2Q2P
 
 | |
2Q39
 
 | |
2Q2M
 
 | |
1LJJ
 
 | |
1LJE
 
 | |
1LJF
 
 | |
