6I7W
 
 | | Structure of the periplasmic binding protein (PBP) AccA in complex with 2-glucose-2-O-lactic acid phosphate (G2LP) from Agrobacterium fabrum C58 | | Descriptor: | 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-beta-D-glucopyranose, ABC transporter, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of a non-natural glucose-2-phosphate ester able to dupe the acc system of Agrobacterium fabrum.
Org. Biomol. Chem., 17, 2019
|
|
3FKB
 
 | | Structure of NDPK H122G and tenofovir-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morera, S, Chen, Y.X. | | Deposit date: | 2008-12-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
1NDP
 
 | | ADENOSINE 5'-DIPHOSPHATE BINDING AND THE ACTIVE SITE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Dumas, C, Lascu, I, Lebras, G, Veron, M. | | Deposit date: | 1993-11-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenosine 5'-diphosphate binding and the active site of nucleoside diphosphate kinase.
Biochemistry, 33, 1994
|
|
1NSP
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Chiadmi, M, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-18 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
1NSQ
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Chiadmi, M, Morera, S, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
3N2S
 
 | | Structure of NfrA1 nitroreductase from B. subtilis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH-dependent nitro/flavin reductase | | Authors: | Morera, S, Gueguen-Chaignon, V, Meyer, P, Cortial, S, Ouazzani, J. | | Deposit date: | 2010-05-19 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role.
Febs Lett., 584, 2010
|
|
8OF3
 
 | | Structure of the apoform of ALDEHYDE DEHYDROGENASE 5F1 (ALDH5F1) from the moss Physcomitrium patens | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
8OFM
 
 | |
8OF1
 
 | | Structure of ALDH5F1 from moss Physcomitrium patens in complex with NAD+ in the contracted conformation | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
3FJO
 
 | | Structure of chimeric YH CPR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Morera, S, Aigrain, L, Truan, G. | | Deposit date: | 2008-12-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase
Embo Rep., 10, 2009
|
|
8RF7
 
 | |
8RGJ
 
 | |
8RPA
 
 | | Crystal structure of Zea mays adenosine kinase 3 (ZmADK3) in complex with AP5A | | Descriptor: | 1,2-ETHANEDIOL, Adenosine kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | A monomer-dimer switch modulates the activity of plant adenosine kinase.
J.Exp.Bot., 2025
|
|
4I8P
 
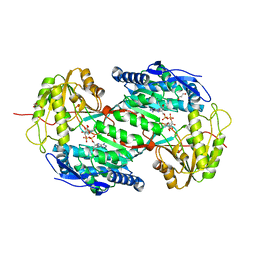 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I8Q
 
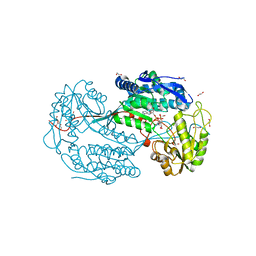 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4I9B
 
 | | Structure of aminoaldehyde dehydrogenase 1 from Solanum lycopersium (SlAMADH1) with a thiohemiacetal intermediate | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
6TG3
 
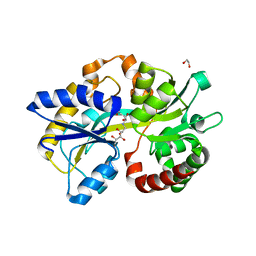 | | Crystal Structure of the PBP/SBP MotA in complex with glucopinic acid from A. tumefaciens B6/R10 | | Descriptor: | (2~{S})-2-[[(3~{S},4~{R},5~{R})-3,4,5,6-tetrakis(oxidanyl)-2-oxidanylidene-hexyl]amino]pentanedioic acid, 1,2-ETHANEDIOL, MotA | | Authors: | Morera, S, Vigouroux, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Import pathways of the mannityl-opines into the bacterial pathogen Agrobacterium tumefaciens: structural, affinity and in vivo approaches.
Biochem.J., 477, 2020
|
|
3IP7
 
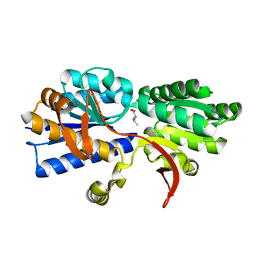 | | Structure of Atu2422-GABA receptor in complex with valine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), CALCIUM ION, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP5
 
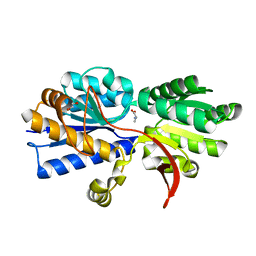 | | Structure of Atu2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP6
 
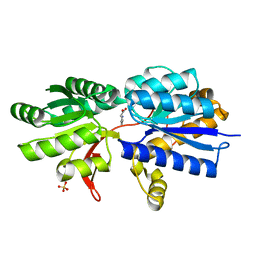 | | Structure of Atu2422-GABA receptor in complex with proline | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), PROLINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPC
 
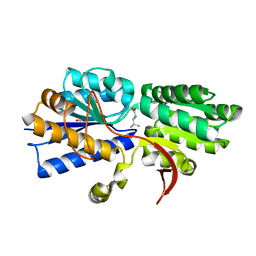 | | Structure of ATU2422-GABA F77A mutant receptor in complex with leucine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), LEUCINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IPA
 
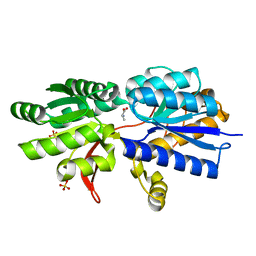 | | Structure of ATU2422-GABA receptor in complex with alanine | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), ALANINE, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
3IP9
 
 | | Structure of Atu2422-GABA receptor in complex with GABA | | Descriptor: | ABC transporter, substrate binding protein (Amino acid), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S, Vigouroux, A. | | Deposit date: | 2009-08-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved mechanism of GABA binding and antagonism is revealed by structure-function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens.
J.Biol.Chem., 285, 2010
|
|
6EPY
 
 | |
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
