2ZE0
 
 | | Alpha-glucosidase GSJ | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Shirai, T, Hung, V.S, Morinaka, K, Kobayashi, T, Ito, S. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GH13 alpha-glucosidase GSJ from one of the deepest sea bacteria
Proteins, 73, 2008
|
|
2ZP1
 
 | |
1WSD
 
 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
2DIE
 
 | | Alkaline alpha-amylase AmyK from Bacillus sp. KSM-1378 | | Descriptor: | CALCIUM ION, SODIUM ION, amylase | | Authors: | Shirai, T, Igarashi, K, Ozawa, T, Hagihara, H, Kobayashi, T, Ozaki, K, Ito, S. | | Deposit date: | 2006-03-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ancestral sequence evolutionary trace and crystal structure analyses of alkaline alpha-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins
Proteins, 66, 2007
|
|
3WVH
 
 | | Time-Resolved Crystal Structure of HindIII with 25sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WVI
 
 | | Time-Resolved Crystal Structure of HindIII with 40 sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WVP
 
 | | Time-Resolved Crystal Structure of HindIII with 60sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*A)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-06-02 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VP7
 
 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
3S6Y
 
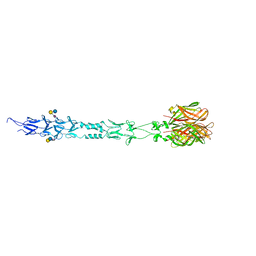 | | Structure of reovirus attachment protein sigma1 in complex with alpha-2,6-sialyllactose | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Outer capsid protein sigma-1 | | Authors: | Reiter, D.M, Dermody, T.S, Stehle, T. | | Deposit date: | 2011-05-26 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides
Plos Pathog., 7, 2011
|
|
3S6X
 
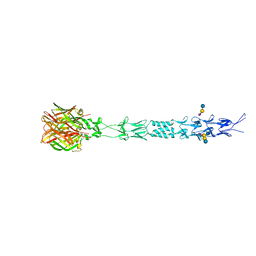 | |
3S6Z
 
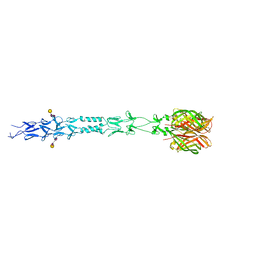 | |
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
3KLR
 
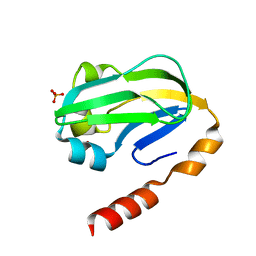 | | Bovine H-protein at 0.88 angstrom resolution | | Descriptor: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | Authors: | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | Deposit date: | 2009-11-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5XJM
 
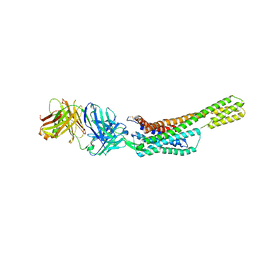 | | Complex structure of angiotensin II type 2 receptor with Fab | | Descriptor: | FabH, FabL, Sar1, ... | | Authors: | Asada, H, Horita, S, Shimamura, T, Iwata, S. | | Deposit date: | 2017-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XLI
 
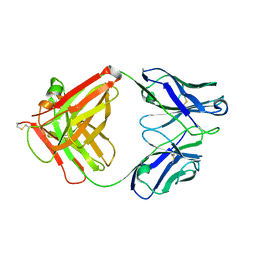 | | Structure of anti-Angiotensin II type2 receptor antibody (D5711-4A03) | | Descriptor: | FabH, FabL | | Authors: | Asada, H, Horita, S, Iwata, S, Hirata, K. | | Deposit date: | 2017-05-10 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5NNW
 
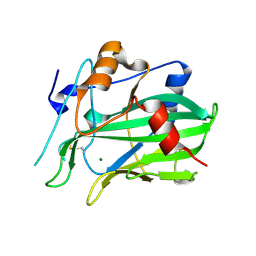 | | NLPPya in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
5NO9
 
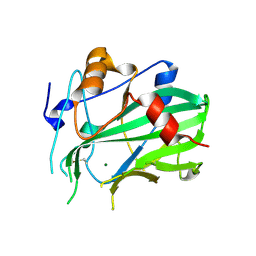 | | NLPPya in complex with mannosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-mannopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
