4N8X
 
 | | The structure of Nostoc sp. PCC 7120 CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Keeling, T.J, Kimber, M.S. | | Deposit date: | 2013-10-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interactions and structural variability of beta-carboxysomal shell protein CcmL.
Photosynth.Res., 121, 2014
|
|
3KWD
 
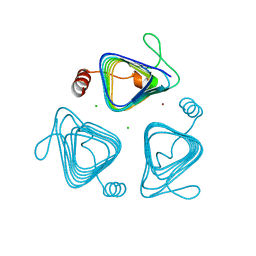 | | Inactive truncation of the beta-carboxysomal gamma-Carbonic Anhydrase, CcmM, form 1 | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, ZINC ION | | Authors: | Pena, K.L, Kimber, M.S, Castel, S.E. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the oxidative activation of the carboxysomal {gamma}-carbonic anhydrase, CcmM.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6CI9
 
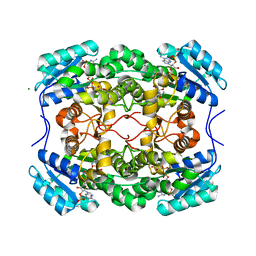 | |
6CI8
 
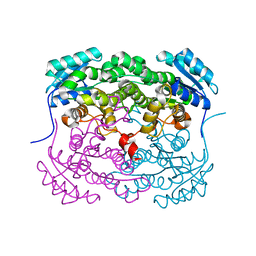 | |
6EF6
 
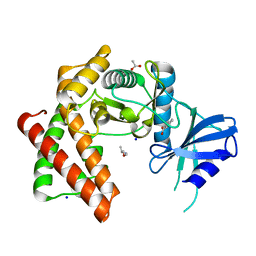 | | Structure of the microcompartment-associated aminopropanol kinase | | Descriptor: | (2R)-1-methoxypropan-2-amine, ACETATE ION, Aminoglycoside phosphotransferase, ... | | Authors: | Mallette, E, Kimber, M.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and kinetic characterization of (S)-1-amino-2-propanol kinase from the aminoacetone utilization microcompartment ofMycobacterium smegmatis.
J.Biol.Chem., 293, 2018
|
|
3GGP
 
 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3SSQ
 
 | | CcmK2 - form 1 dodecamer | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein, GLYCEROL | | Authors: | Samborska, B, Kimber, M.S. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A CcmK2 double layer is the dominant architectural feature of the beta-carboxysomal shell facet
Structure, 2012
|
|
5SUH
 
 | |
3Q9O
 
 | |
2Q6M
 
 | |
2Q5T
 
 | | Full-length Cholix toxin from Vibrio Cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cholix toxin | | Authors: | Jorgensen, R, Fieldhouse, R.J, Merrill, A.R. | | Deposit date: | 2007-06-01 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cholix Toxin, a Novel ADP-ribosylating Factor from Vibrio cholerae.
J.Biol.Chem., 283, 2008
|
|
3GGO
 
 | | Crystal structure of prephenate dehydrogenase from A. aeolicus with HPP and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Prephenate dehydrogenase | | Authors: | Sun, W, Shahinas, D, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
3GGG
 
 | |
4FXQ
 
 | | Full-length Certhrax toxin from Bacillus cereus in complex with Inhibitor P6 | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, CHLORIDE ION, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Visschedyk, D.D, Dimov, S, Kimber, M.S, Park, H.W, Merrill, A.R. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9599 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4FML
 
 | |
2NWQ
 
 | | Short chain dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Probable short-chain dehydrogenase, SULFATE ION | | Authors: | McGrath, T.E, Kimber, M.S, Beattie, B, Thambipillai, D, Dharamsi, A, Virag, C, Nethery-Brokx, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short chain dehydrogenase from Pseudomonas aeruginosa
To be Published
|
|
