2JZZ
 
 | |
2NSW
 
 | |
2NSV
 
 | |
2KWU
 
 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWV
 
 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KA7
 
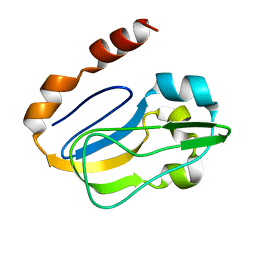 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
2K9Z
 
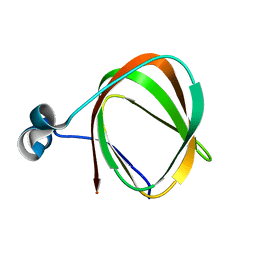 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA0
 
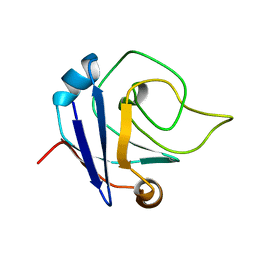 | | NMR structure of the protein TM1367 | | Descriptor: | uncharacterized protein TM1367 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KL2
 
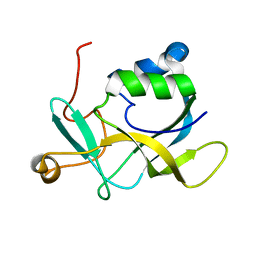 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2LU5
 
 | | Structure and chemical shifts of Cu(I),Zn(II) superoxide dismutase by solid-state NMR | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Knight, M.J, Pell, A.J, Bertini, I, Felli, I.C, Gonnelli, L, Pierattelli, R, Herrmann, T, Emsley, L, Pintacuda, G. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JNQ
 
 | | Solution Structure of a KlbA Intein Precursor from Methanococcus jannaschii | | Descriptor: | Hypothetical protein MJ0781 | | Authors: | Johnson, M.A, Southworth, M.W, Herrmann, T, Brace, L, Perler, F.B, Wuthrich, K.A. | | Deposit date: | 2007-01-31 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a KlbA intein precursor from Methanococcus jannaschii
Protein Sci., 16, 2007
|
|
2JMZ
 
 | | Solution structure of a KlbA intein precursor from Methanococcus jannaschii | | Descriptor: | Hypothetical protein MJ0781 | | Authors: | Johnson, M.A, Southworth, M.W, Herrmann, T, Brace, L, Perler, F.B, Wuthrich, K.A. | | Deposit date: | 2006-12-14 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a KlbA intein precursor from Methanococcus jannaschii
Protein Sci., 16, 2007
|
|
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | Descriptor: | fimF | | Authors: | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
6QH7
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6UJV
 
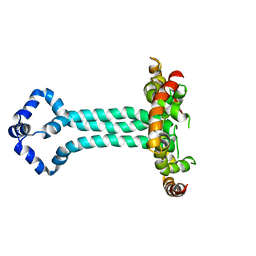 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
5JYN
 
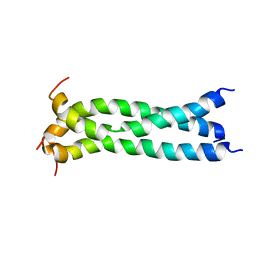 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
6QH5
 
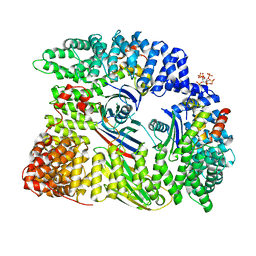 | | AP2 clathrin adaptor mu2T156-phosphorylated core in closed conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH6
 
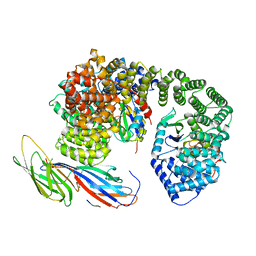 | | AP2 clathrin adaptor core with two cargo peptides in open+ conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
2QJB
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant IA/IB | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA, CHLORIDE ION | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2QJ9
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B1 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2QJA
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B12 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
1XYU
 
 | | Solution structure of the sheep prion protein with polymorphism H168 | | Descriptor: | Major prion protein | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2LLZ
 
 | | GhoS (YjdK) monomer | | Descriptor: | Uncharacterized protein yjdK | | Authors: | Lord, D, Peti, W, Page, R. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS.
Nat.Chem.Biol., 8, 2012
|
|
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2JPO
 
 | |
