7C5R
 
 | | Crystal Structure of C150S mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.31 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5Q
 
 | | Crystal Structure of H177A mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.13 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5I
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 2.49 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5K
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.69 Angstrom resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5P
 
 | | Crystal Structure Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.35 Angstrom resolution. | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5L
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5J
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C7K
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from Escherichia coli at 1.77 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5O
 
 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5M
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 1.8 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
5D6J
 
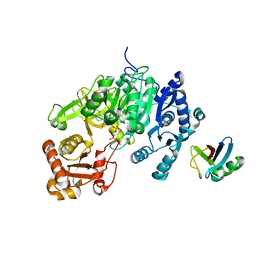 | | Crystal structure of a mycobacterial protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-CoA synthase, MAGNESIUM ION, ... | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
8X5D
 
 | |
5D6N
 
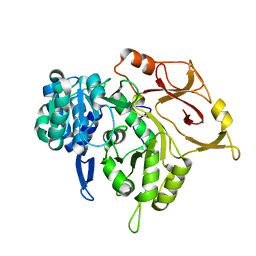 | | Crystal structure of a mycobacterial protein | | Descriptor: | Acyl-CoA synthase | | Authors: | Li, W.J, Bi, L.J. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of FadD32, an enzyme essential for mycolic acid biosynthesis in mycobacteria.
Sci Rep, 5, 2015
|
|
6KBD
 
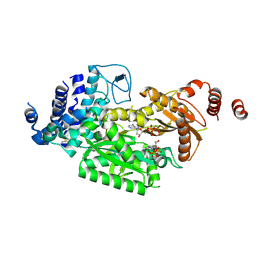 | | fused To-MtbCsm1 with 2dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
7R97
 
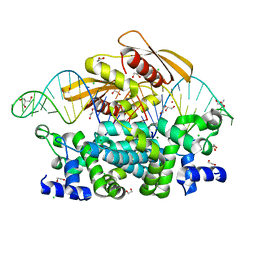 | | Crystal structure of postcleavge complex of Escherichia coli RNase III | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Dharavath, S, Shaw, G.X, Ji, X. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg 2+ -ion catalysis.
Rna Biol., 19, 2022
|
|
6KC0
 
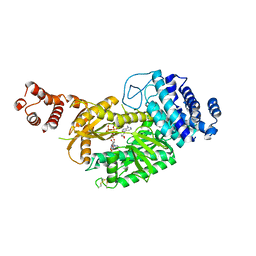 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
4HCC
 
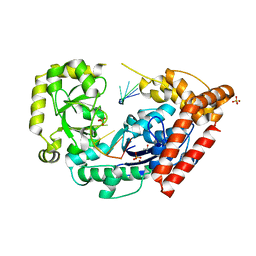 | | The zinc ion bound form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, ISOPROPYL ALCOHOL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
4HCB
 
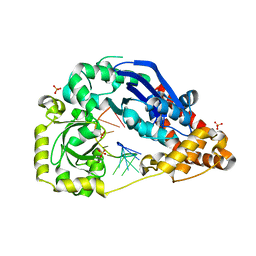 | | The metal-free form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, GLYCEROL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
3V65
 
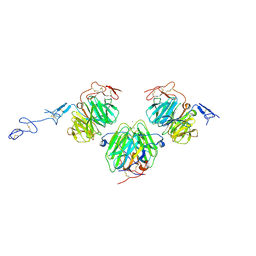 | | Crystal structure of agrin and LRP4 complex | | Descriptor: | Agrin, CALCIUM ION, Low-density lipoprotein receptor-related protein 4 | | Authors: | Zong, Y, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
7F1N
 
 | | Beta-Glucosidase | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Anke, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14000368 Å) | | Cite: | Structural and Catalytic Characterization of TsBGL, a beta-Glucosidase From Thermofilum sp. ex4484_79.
Front Microbiol, 12, 2021
|
|
