5G33
 
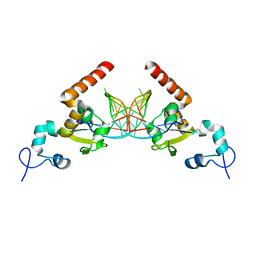 | | Structure of Rad14 in complex with acetylnaphtyl-guanine containing DNA | | Descriptor: | 5'-D(*GP*CP*TP*CP*TP*AP*MFOP*TP*CP*AP*TP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*GP)-3', RAD14, ... | | Authors: | Simon, N, Ebert, C, Schneider, S. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Bulky Adduct DNA Lesion Recognition by the Nucleotide Excision Repair Protein Rad14.
Chemistry, 22, 2016
|
|
6S8Z
 
 | |
1EM0
 
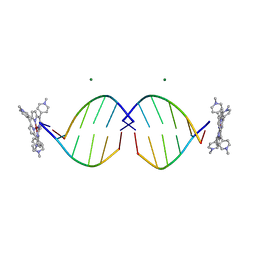 | | COMPLEX OF D(CCTAGG) WITH TETRA-[N-METHYL-PYRIDYL] PORPHYRIN | | Descriptor: | DNA (5'-D(*(CBR)P*CP*TP*AP*GP*G)-3'), MAGNESIUM ION, TETRA[N-METHYL-PYRIDYL] PORPHYRIN-NICKEL | | Authors: | Neidle, S, Sanderson, M, Bennett, M, Krah, A, Wien, F, Garman, E, McKenna, R. | | Deposit date: | 2000-03-14 | | Release date: | 2000-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2GVR
 
 | |
7AYX
 
 | |
4CIS
 
 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4ELT
 
 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5-[(4-ethynylphenyl)ethynyl]uridine 5'-(tetrahydrogen triphosphate), ACETATE ION, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Chem.Commun.(Camb.), 48, 2012
|
|
4ELV
 
 | |
4ELU
 
 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines | | Descriptor: | 2'-deoxy-5-[(4-ethynylphenyl)ethynyl]cytidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Chem.Commun.(Camb.), 48, 2012
|
|
3RRG
 
 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddGTP | | Descriptor: | (5'-D(*AP*AP*AP*(3DR)P*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-04-29 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino Acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
3RR8
 
 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddGTP | | Descriptor: | (5'-D(*AP*AP*AP*(3DR)P*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-04-29 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Amino Acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
3RRH
 
 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and a ddTTP | | Descriptor: | (5'-D(*AP*AP*AP*(3DR)P*AP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DT))-3'), ACETATE ION, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-04-29 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amino Acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
3T3F
 
 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and dNITP | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, 5'-D(*AP*AP*AP*(3DR)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3', ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
3RR7
 
 | | Binary Structure of the large fragment of Taq DNA polymerase bound to an abasic site | | Descriptor: | (5'-D(*AP*AP*AP*(3DR)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-04-29 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amino Acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
4CA4
 
 | | Crystal structure of FimH lectin domain with the Tyr48Ala mutation, in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FIMH, heptyl alpha-D-mannopyranoside | | Authors: | Rabbani, S, Bouckaert, J, Zalewski, A, Preston, R, Eid, S, Thompson, A, Puorger, C, Glockshuber, R, Ernst, B. | | Deposit date: | 2013-10-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Mutation of Tyr137 of the universal Escherichia coli fimbrial adhesin FimH relaxes the tyrosine gate prior to mannose binding.
IUCrJ, 4, 2017
|
|
6QF5
 
 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
4B9U
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. | | Descriptor: | 5'-D(*CP*AP*AP*FOXP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
5AC2
 
 | | human aldehyde dehydrogenase 1A1 with duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, RETINAL DEHYDROGENASE 1, YTTERBIUM (III) ION, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4B9T
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dC basepair in the post-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9M
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dA DNA lesion -thymine basepair in the post- insertion site. | | Descriptor: | 5'-D(*DC*DA*DA*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
5ABM
 
 | | Sheep aldehyde dehydrogenase 1A1 | | Descriptor: | MAGNESIUM ION, RETINAL DEHYDROGENASE 1, [[(2R,3S,4R,5R)-5-[(3R)-3-aminocarbonyl-3,4-dihydro-2H-pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanidyl-ph osphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7P98
 
 | | Cyclohex-1-ene-1-carboxyl-CoA dehydrogenase in a substrate-free state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Short-chain acyl-CoA dehydrogenase | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
7P9X
 
 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1-ene-1-carboxyl-CoA | | Descriptor: | 1-monoenoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
7P9A
 
 | | Structure of cyclohex-1-ene-1-carboxyl-CoA dehydrogenase complexed with cyclohex-1,5-diene-1-carboxyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ermler, U, Weidenweber, S, Boll, M. | | Deposit date: | 2021-07-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases.
Chembiochem, 22, 2021
|
|
2AVH
 
 | | G4T3G4 dimeric quadruplex structure | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*TP*TP*GP*GP*GP*G)-3', POTASSIUM ION | | Authors: | Hazel, P, Parkinson, G.N, Neidle, S. | | Deposit date: | 2005-08-30 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex.
J.Am.Chem.Soc., 128, 2006
|
|
