8PX9
 
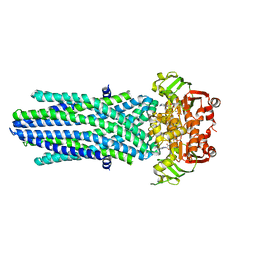 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXC
 
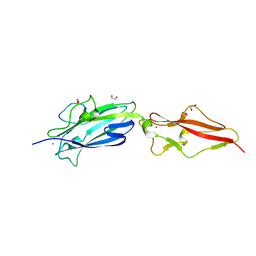 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PWN
 
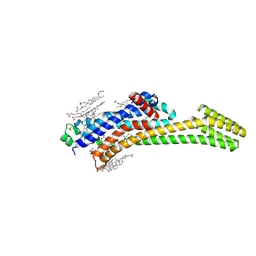 | | Structure of A2A adenosine receptor A2AR-StaR2-bRIL, solved at wavelength 2.75 A | | Descriptor: | Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, OLEIC ACID, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Moraes, I, Wagner, A. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXK
 
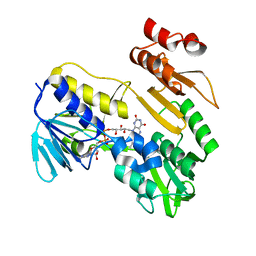 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX1
 
 | | Structure of salmonella effector SseK3, solved at wavelength 2.75 A | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C.M, Esposito, D, Rittinger, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX7
 
 | | Structure of Bacterial Multidrug Efflux transporter AcrB, solved at wavelength 3.02 A | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX5
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX4
 
 | | Structure of the PAS domain code by the LIC_11128 gene from Leptospira interrogans serovar Copenhageni Fiocruz, solved at wavelength 3.09 A | | Descriptor: | Diguanylate cyclase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Guzzo, C.R, Owens, R.J, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXJ
 
 | | Structure of Whitewater Arroyo virus GP1 glycoprotein, solved at wavelength 2.75 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Glycoprotein G1, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bowden, T.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX0
 
 | | Structure of ribonuclease A, solved at wavelength 2.75 A | | Descriptor: | L-URIDINE-5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXG
 
 | | Structure of Streptactin, solved at wavelength 2.75 A | | Descriptor: | CHLORIDE ION, GLYCEROL, Streptavidin | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Vecchia, L, Jones, E.Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXL
 
 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 1.37 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXH
 
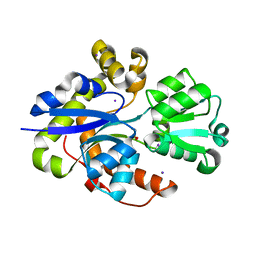 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
3ZIH
 
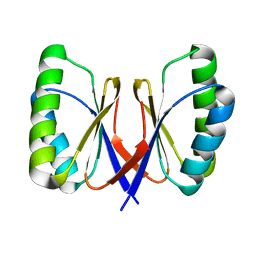 | | Bacillus subtilis SepF, C-terminal domain | | Descriptor: | CELL DIVISION PROTEIN SEPF | | Authors: | Duman, R.E, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CJ7
 
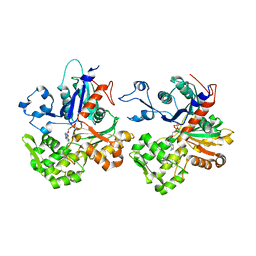 | | Structure of Crenactin, an archeal actin-like protein | | Descriptor: | ACTIN/ACTIN FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Izore, T, Duman, R.E, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crenactin from Pyrobaculum Calidifontis is Closely Related to Actin in Structure and Forms Steep Helical Filaments.
FEBS Lett., 588, 2014
|
|
7PEO
 
 | | Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, HOLMIUM ATOM, ... | | Authors: | von Kugelgen, A, Bharat, T.A.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | High-resolution mapping of metal ions reveals principles of surface layer assembly in Caulobacter crescentus cells.
Structure, 30, 2022
|
|
5LQ4
 
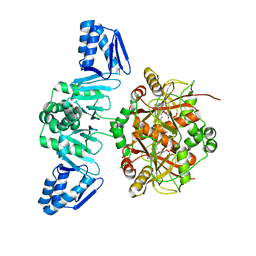 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | Descriptor: | CyaGox, FLAVIN MONONUCLEOTIDE | | Authors: | Bent, A.F, Wagner, A, Naismith, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
9FGP
 
 | |
6EQH
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup C2221 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQD
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQE
 
 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQF
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P212121 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9F56
 
 | |
9F5B
 
 | | Identification of zinc ions in LMO4. | | Descriptor: | LIM domain transcription factor LMO4,LIM domain-binding protein 1, ZINC ION | | Authors: | El Omari, K, Forsyth, I, Mancini, E.J, Wagner, A. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Utilizing anomalous signals for element identification in macromolecular crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
