8QDZ
 
 | |
8QE1
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 15 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-6-(4-methoxyphenyl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE3
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 31 | | Descriptor: | 3-cyclopropyl-6-(2-methylindazol-5-yl)-4-(6-methylpyridin-3-yl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QDY
 
 | |
5FPO
 
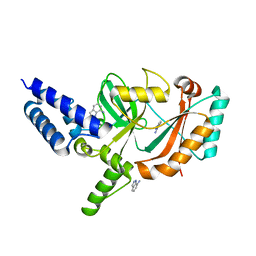 | | Structure of Bacterial DNA Ligase with small-molecule ligand 1H- indazol-7-amine (AT4213) in an alternate binding site. | | Descriptor: | 1H-indazol-7-amine, DNA LIGASE | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPR
 
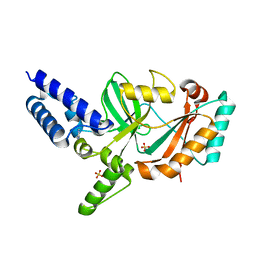 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
