3ZUU
 
 | | The structure of OST1 (D160A, S175D) kinase in complex with gold | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Serine/threonine-protein kinase SRK2E | | Authors: | Yunta, C, Martinez-Ripoll, M, Albert, A. | | Deposit date: | 2011-07-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress.
J. Mol. Biol., 414, 2011
|
|
3ZML
 
 | |
3ZMK
 
 | |
3ZUT
 
 | | The structure of OST1 (D160A) kinase | | Descriptor: | Serine/threonine-protein kinase SRK2E | | Authors: | Yunta, C, Martinez-Ripoll, M, Albert, A. | | Deposit date: | 2011-07-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress.
J. Mol. Biol., 414, 2011
|
|
1MVN
 
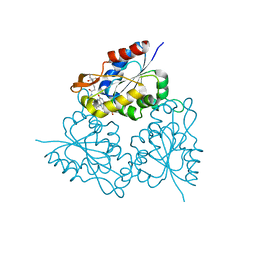 | | PPC decarboxylase mutant C175S complexed with pantothenoylaminoethenethiol | | Descriptor: | 2,4-DIHYDROXY-N-[2-(2-MERCAPTO-VINYLCARBAMOYL)-ETHYL]-3,3-DIMETHYL-BUTYRAMIDE, FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
4RGD
 
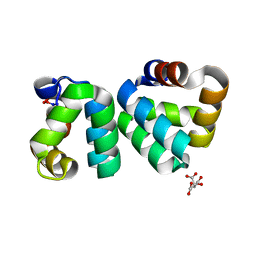 | | The structure a AS-48 G13K/L40K mutant | | Descriptor: | Bacteriocin AS-48, CITRIC ACID | | Authors: | Sanchez-Barrena, M.J. | | Deposit date: | 2014-09-29 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The bacteriocin AS-48 requires dimer dissociation followed by hydrophobic interactions with the membrane for antibacterial activity.
J.Struct.Biol., 190, 2015
|
|
6Q5Z
 
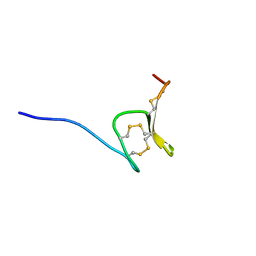 | | H-Vc7.2, H-superfamily conotoxin | | Descriptor: | Conotoxin Vc7.2 | | Authors: | Nielsen, L.D, Foged, M.M, Teilum, K, Ellgaard, L. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework.
J.Biol.Chem., 294, 2019
|
|
2IXV
 
 | |
5A52
 
 | |
2J8G
 
 | |
2J8F
 
 | |
2IXU
 
 | |
2EHB
 
 | |
5MMX
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CSPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Moreno-Alvero, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MMQ
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | CSPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
3UTL
 
 | | Human pepsin 3b | | Descriptor: | Pepsin A | | Authors: | Coker, A, Cooper, J.B. | | Deposit date: | 2011-11-25 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3URJ
 
 | | Type IV native endothiapepsin | | Descriptor: | Endothiapepsin, SULFATE ION | | Authors: | Bailey, D, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2C45
 
 | |
1MVL
 
 | | PPC decarboxylase mutant C175S | | Descriptor: | FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
1PQF
 
 | | Glycine 24 to Serine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYQ
 
 | | Unprocessed Aspartate Decarboxylase Mutant, with Alanine inserted at position 24 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PT1
 
 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with Histidine 11 Mutated to Alanine | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PQH
 
 | | Serine 25 to Threonine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, MALONIC ACID, SODIUM ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PPY
 
 | | Native precursor of pyruvoyl dependent Aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase precursor, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYU
 
 | | Processed Aspartate Decarboxylase Mutant with Ser25 mutated to Cys | | Descriptor: | Aspartate 1-decarboxylase alfa chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
