1B2B
 
 | |
1B2E
 
 | |
1B17
 
 | |
1B2G
 
 | |
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1B2A
 
 | |
1B18
 
 | |
4C69
 
 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1C61
 
 | |
1C60
 
 | |
1C62
 
 | |
1C65
 
 | |
1C64
 
 | |
1C6E
 
 | |
1C69
 
 | |
1C6B
 
 | |
1C63
 
 | |
1C6C
 
 | |
1C6D
 
 | |
1C6F
 
 | |
1C6H
 
 | |
1C6G
 
 | |
3OQ1
 
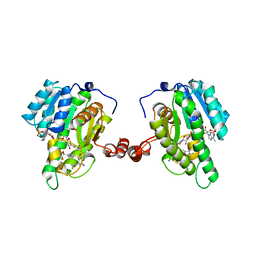 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Diarylsulfone Inhibitor | | Descriptor: | 3-(2-fluoroethyl)-4-({4-[(2S)-1,1,1-trifluoro-2-hydroxypropan-2-yl]phenyl}sulfonyl)benzonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The synthesis and SAR of novel diarylsulfone 11beta-HSD1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1C6I
 
 | |
