1IUK
 
 | | The structure of native ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
8Y98
 
 | | Crystal structure of a heterooligomeric aminotransferase from Serratia sp. ATCC 39006, PPE-bound form | | Descriptor: | DegT/DnrJ/EryC1/StrS aminotransferase, DegT/DnrJ/EryC1/StrS family aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Pramono, H, Yoshida, A, Nishiyama, M. | | Deposit date: | 2024-02-06 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of a novel heterooligomeric aminotransferase from Serratia sp. ATCC 39006 provides insights into function.
Febs Lett., 599, 2025
|
|
8Y96
 
 | |
8Y97
 
 | | Crystal structure of a heterooligomeric aminotransferase from Serratia sp. ATCC 39006, PMP-bound form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DegT/DnrJ/EryC1/StrS aminotransferase, DegT/DnrJ/EryC1/StrS family aminotransferase | | Authors: | Pramono, H, Yoshida, A, Nishiyama, M. | | Deposit date: | 2024-02-06 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a novel heterooligomeric aminotransferase from Serratia sp. ATCC 39006 provides insights into function.
Febs Lett., 599, 2025
|
|
8ZTZ
 
 | | Structure of ATP-dependent diazotase CmaA6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Putative AMP-binding enzyme | | Authors: | Katsuyama, Y, Kawai, S, Ohnishi, Y. | | Deposit date: | 2024-06-07 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of ATP-Dependent Diazotase CmaA6.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7V1W
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-arabinofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase (alphaFFase1), beta-D-arabinofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1X
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-fructofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase, beta-D-fructofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
1UDX
 
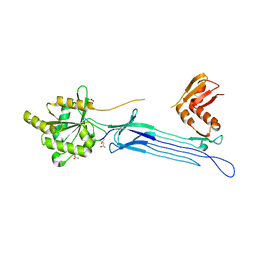 | | Crystal structure of the conserved protein TT1381 from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, the GTP-binding protein Obg | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Murayama, K, Inoue, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-07 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8.
J.Mol.Biol., 337, 2004
|
|
1WIJ
 
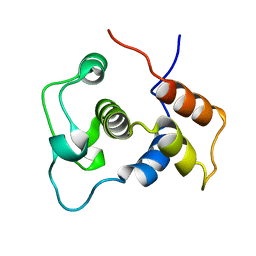 | | Solution Structure of the DNA-Binding Domain of Ethylene-Insensitive3-Like3 | | Descriptor: | ETHYLENE-INSENSITIVE3-like 3 protein | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3.
J.Mol.Biol., 348, 2005
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | Descriptor: | Probable WRKY transcription factor 4, ZINC ION | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
1WID
 
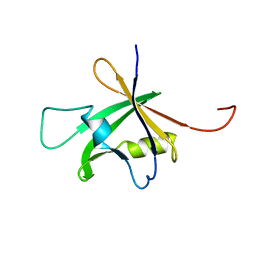 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
1UL5
 
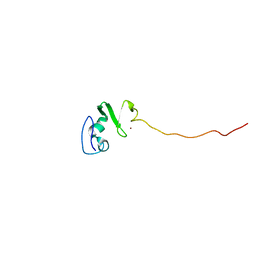 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 7 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
1UL4
 
 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 4 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
2RQR
 
 | | The solution structure of human DOCK2 SH3 domain - ELMO1 peptide chimera complex | | Descriptor: | Engulfment and cell motility protein 1,Dedicator of cytokinesis protein 2 | | Authors: | Yokoyama, S, Tochio, N, Koshiba, S, Kigawa, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-21 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1WJ0
 
 | |
1UL7
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
1WVO
 
 | | Solution structure of RSGI RUH-029, an antifreeze protein like domain in human N-acetylneuraminic acid phosphate synthase gene. | | Descriptor: | Sialic acid synthase | | Authors: | Ito, Y, Hamada, T, Hayashi, F, Yokoyama, S, Hirota, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-22 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antifreeze-like domain of human sialic acid synthase
Protein Sci., 15, 2006
|
|
3VJN
 
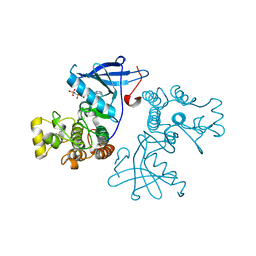 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
1WMG
 
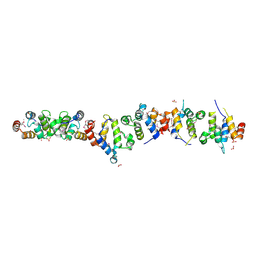 | | Crystal structure of the UNC5H2 death domain | | Descriptor: | SULFATE ION, SULFITE ION, netrin receptor Unc5h2 | | Authors: | Handa, N, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-09 | | Release date: | 2005-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the UNC5H2 death domain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1WWQ
 
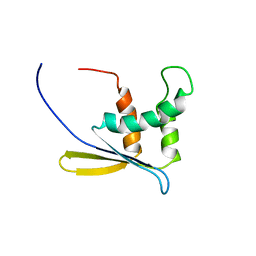 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
7VN7
 
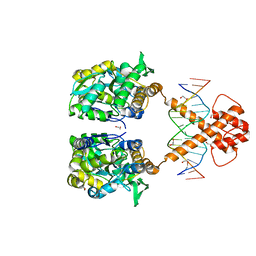 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GACACGTGTC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*AP*CP*AP*CP*GP*TP*GP*TP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN8
 
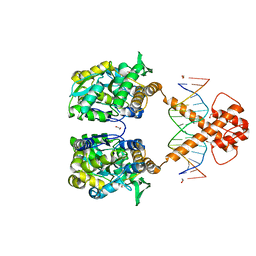 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning GTCACGTGAC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN3
 
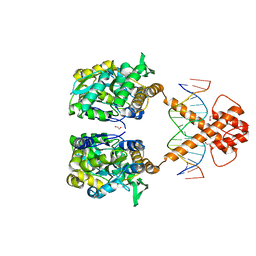 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CACACGTGTG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
