6ZRC
 
 | |
7AKP
 
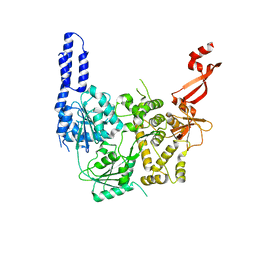 | | Crystal structure of E. coli RNA helicase HrpA-D305A | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2IJE
 
 | |
1L6U
 
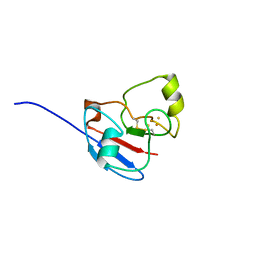 | | NMR STRUCTURE OF OXIDIZED ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
7R0N
 
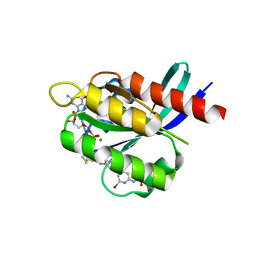 | | KRasG12C in complex with GDP and compound 2 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0M
 
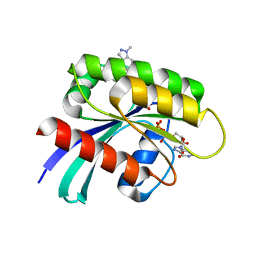 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0Q
 
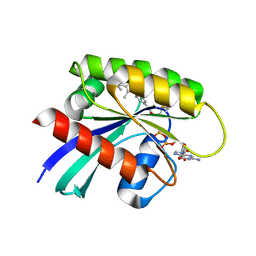 | | KRasG12C in complex with GDP and compound 3 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
6YQG
 
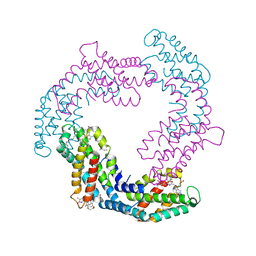 | | Crystal structure of native Phycocyanin in spacegroup P63 at 1.45 Angstroms. | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1YJJ
 
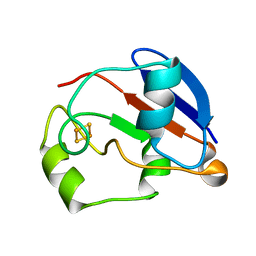 | | RDC-refined Solution NMR structure of oxidized putidaredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Jain, N.U, Tjioe, E, Savidor, A, Boulie, J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings.
Biochemistry, 44, 2005
|
|
6Y6O
 
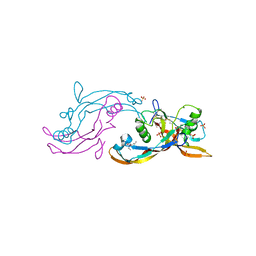 | | Structure of mature activin A with small molecule 42 | | Descriptor: | (3~{R})-4-ethyl-3-methyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6Y6N
 
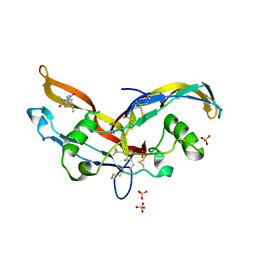 | | Structure of mature activin A with small molecule 2 | | Descriptor: | (3~{R})-3,4-dimethyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6YPQ
 
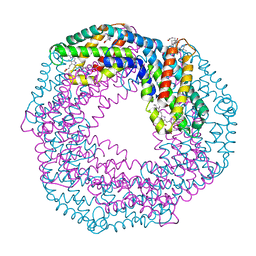 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup R32 at 1.29 Angstroms | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCINE, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4WNX
 
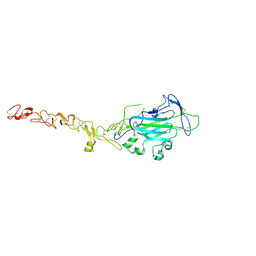 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
6YQ8
 
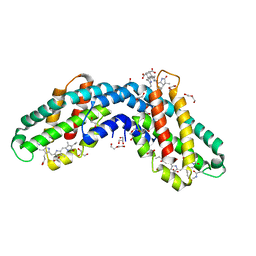 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P63 at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7C3N
 
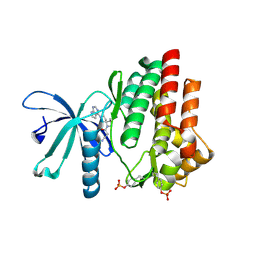 | | Crystal structure of JAK3 in complex with Delgocitinib | | Descriptor: | 3-[(3S,4R)-3-methyl-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,7-diazaspiro[3.4]octan-1-yl]-3-oxidanylidene-propanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Doi, S, Otira, T, Kikuwaka, M, Nomura, A, Noji, S, Adachi, T. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
J.Med.Chem., 63, 2020
|
|
8J2P
 
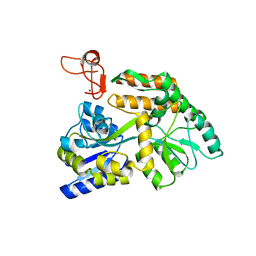 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
7C3A
 
 | | Ferredoxin reductase in carbazole 1,9a-dioxygenase | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ashikawa, Y, Fujimoto, Z, Nojiri, H. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ferredoxin reductase component of carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7C3B
 
 | | Ferredoxin reductase in carbazole 1,9a-dioxygenase (FAD apo form) | | Descriptor: | ACETATE ION, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Fujimoto, Z, Nojiri, H. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ferredoxin reductase component of carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8PCA
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 50 ms | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCP
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 250 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCD
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 150 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCM
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 80 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCR
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 750 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCE
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 250 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
