2ZE1
 
 | | X-ray structure of Bace-1 in complex with compound 6g | | Descriptor: | 3-bromo-N-[4-[1-(2-carbamimidamido-2-oxo-ethyl)-5-phenyl-pyrrol-2-yl]phenyl]benzamide, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7C7P
 
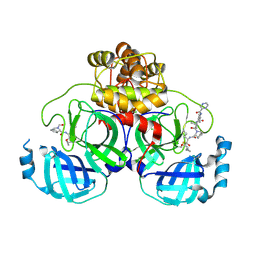 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7C8D
 
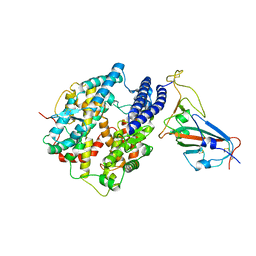 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7W8S
 
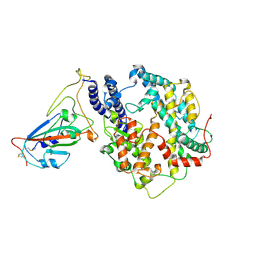 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
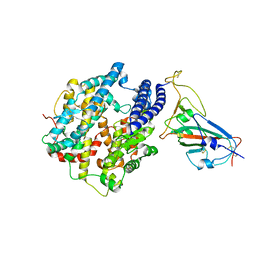 | |
7WSF
 
 | |
7WSE
 
 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
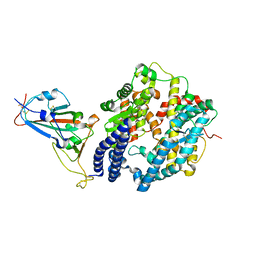 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
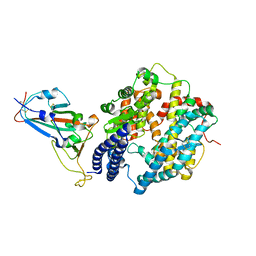 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
8XA2
 
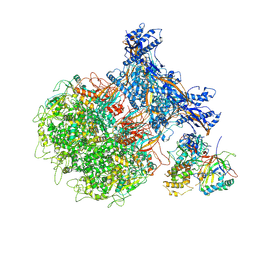 | | Penton capsomer of the VZV B-Capsid | | Descriptor: | Major capsid protein, Tri1, Tri2A, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8XA0
 
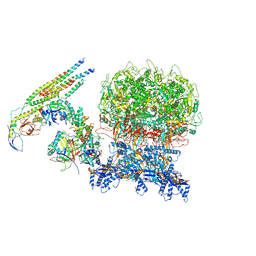 | | penton capsomer of the VZV C-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8XA1
 
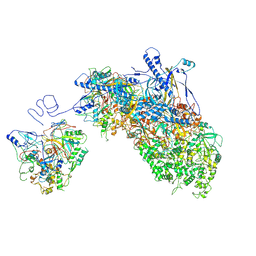 | | Portal vertex capsomer of VZV B-capsid | | Descriptor: | Major capsid protein, Tri1, Tri2A, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9W
 
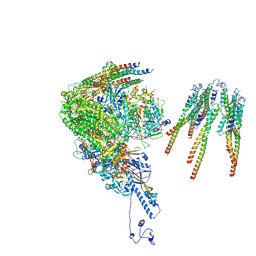 | | portal vertex capsomer of the VZV C-Capsid | | Descriptor: | CVC1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Nan, W, Lei, C, Jiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8XA3
 
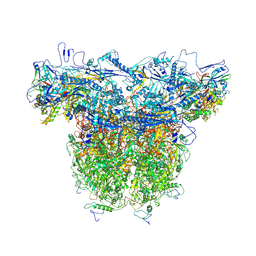 | | C-hexon capsomer of the VZV B-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Wang, N, Cao, L, Wang, X. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9X
 
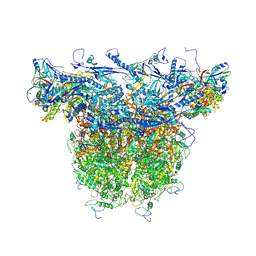 | | C-hexon capsomer of the VZV C-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9Y
 
 | | E-hexon capsomer of the VZV C-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8XSD
 
 | | BA.5 Spike complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike glycoprotein | | Authors: | Feng, L.L. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
8Z86
 
 | | BA.5 RBD in complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike protein S1 | | Authors: | Feng, L.L. | | Deposit date: | 2024-04-21 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
