6KCR
 
 | |
6L3T
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the absence of calcium | | Descriptor: | Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L3V
 
 | | The R15G mutant of human Cx31.3/GJC3 connexin hemichannel | | Descriptor: | Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L3U
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the presence of calcium | | Descriptor: | CALCIUM ION, Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
5B16
 
 | | X-ray structure of DROSHA in complex with the C-terminal tail of DGCR8. | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3,DROSHA,Ribonuclease 3,DROSHA,Ribonuclease 3, ZINC ION | | Authors: | Kwon, S.C, Nguyen, T.A, Choi, Y.G, Jo, M.H, Hohng, S, Kim, V.N, Woo, J.S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human DROSHA
Cell, 164, 2016
|
|
4K1V
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y57F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
4K1U
 
 | | Crystal structure of delta5-3-ketosteroid isomerase containing Y16F and Y32F mutations | | Descriptor: | Steroid Delta-isomerase | | Authors: | Cha, H.J, Jang, D.S, Kim, Y.G, Hong, B.H, Woo, J.S, Choi, K.Y. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase
Mol.Cells, 36, 2013
|
|
2WP7
 
 | | Crystal structure of deSUMOylase(DUF862) | | Descriptor: | PPPDE PEPTIDASE DOMAIN-CONTAINING PROTEIN 2 | | Authors: | Kim, J.H, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Desi-1, a Novel Desumoylase Belonging to a Putative Isopeptidase Superfamily.
Proteins, 80, 2012
|
|
3MD9
 
 | | Structure of apo form of a periplasmic heme binding protein | | Descriptor: | BROMIDE ION, GLYCEROL, Hemin-binding periplasmic protein hmuT, ... | | Authors: | Mattle, D, Goetz, B.A, Locher, K.P. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
7EP6
 
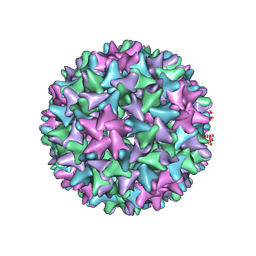 | | Engineered Hepatitis B virus core antigen T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EOY
 
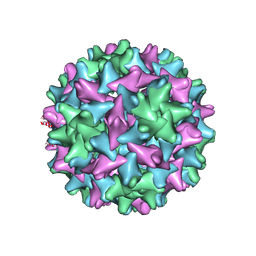 | | Engineered Hepatitis B virus core antigen T=3 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-24 | | Release date: | 2021-09-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7FDJ
 
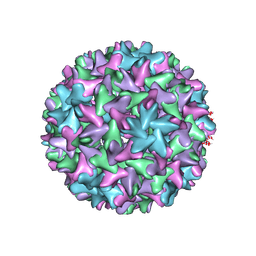 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EPP
 
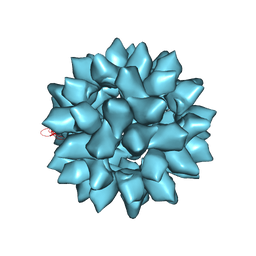 | |
4ZRQ
 
 | |
4ZRP
 
 | | TC:CD320 | | Descriptor: | CALCIUM ION, CD320 antigen, CYANOCOBALAMIN, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2015-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of transcobalamin recognition by human CD320 receptor.
Nat Commun, 7, 2016
|
|
