1MR7
 
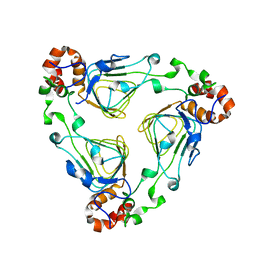 | | Crystal Structure of Streptogramin A Acetyltransferase | | Descriptor: | Streptogramin A Acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
4N1B
 
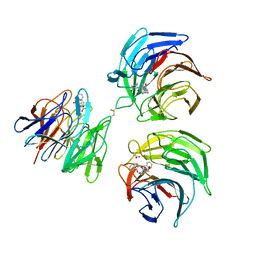 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4P8V
 
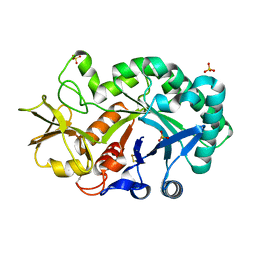 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc2) were solved to resolutions of 1.5 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8W
 
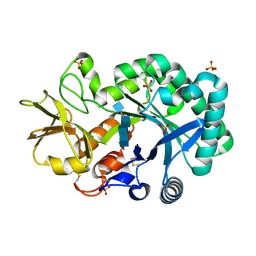 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8X
 
 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc6) were solved to resolutions of 2.48 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8U
 
 | | The crystal structures of YKL-39 in the absence of chitooligosaccharides was solved to resolutions of 2.4 angstrom | | Descriptor: | Chitinase-3-like protein 2, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
5YPU
 
 | | Crystal structure of an actin monomer in complex with the nucleator Cordon-Bleu MET72NLE WH2-motif peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Scipion, C.P.M, Wongsantichon, J, Ferrer, F.J, Yuen, T.Y, Robinson, R.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the roles of divalent cations in actin polymerization and activation of ATP hydrolysis
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YVN
 
 | | Human Glutathione Transferase Omega1 | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase omega-1, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5YVO
 
 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | Descriptor: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
4L7D
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4L7C
 
 | | Structure of keap1 kelch domain with 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione | | Descriptor: | 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4L7B
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4XCM
 
 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9C1R
 
 | | Crystal structure of mutant cMET D1228N kinase domain in complex with inhibitor compound 13 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[(1s,3S)-3-(1-methyl-1H-pyrazol-3-yl)cyclobutyl][(8R)-pyrazolo[1,5-a]pyrazin-4-yl]amino}phenyl)-2-(5-fluoropyridin-2-yl)-3-oxo-2,3-dihydropyridazine-4-carboxamide | | Authors: | Simpson, H, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-29 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Pyrazolopyrazines as Selective, Potent, and Mutant-Active MET Inhibitors with Intracranial Efficacy.
J.Med.Chem., 67, 2024
|
|
8GU0
 
 | | Crystal structure of a fungal halogenase RadH | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Non-heme halogenase radH, ... | | Authors: | Jiang, S.M, Brown, C.J. | | Deposit date: | 2022-09-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Further Characterization of Fungal Halogenase RadH and Its Homologs.
Biomolecules, 13, 2023
|
|
4QPO
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4QPQ
 
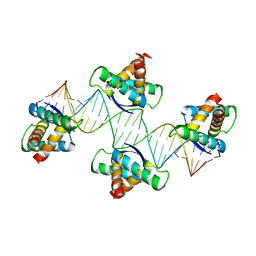 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | Relaxosome protein TraM, sbmA DNA1, sbmA DNA2 | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
6LHM
 
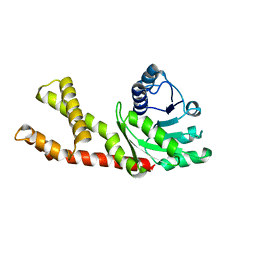 | | Structure of human PYCR2 | | Descriptor: | Pyrroline-5-carboxylate reductase 2 | | Authors: | Baburajendran, N. | | Deposit date: | 2019-12-09 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Loss of PYCR2 Causes Neurodegeneration by Increasing Cerebral Glycine Levels via SHMT2.
Neuron, 107, 2020
|
|
6RCP
 
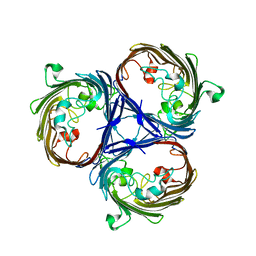 | |
6RD3
 
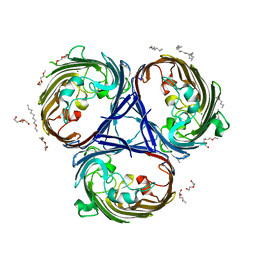 | |
6RCK
 
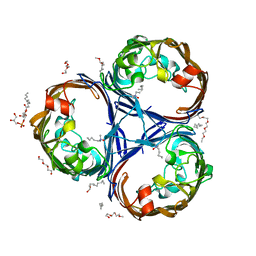 | | Crystal structure of the OmpK36 GD insertion chimera from Klebsiella pneumonia | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Beis, K, Romano, M, Kwong, J. | | Deposit date: | 2019-04-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo.
Nat Commun, 10, 2019
|
|
9FV4
 
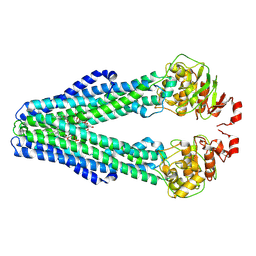 | | MsbA in peptidisc inward-facing narrow (-) open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUQ
 
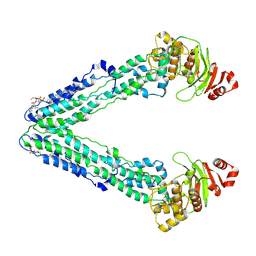 | | MsbA in DDM inward-facing wide open | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV5
 
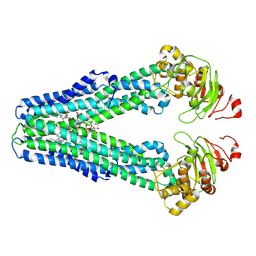 | | MsbA in peptidisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV1
 
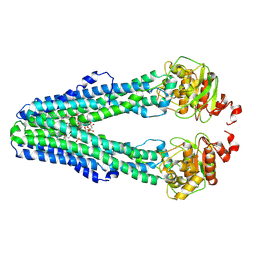 | | MsbA in MSP1D1 (EPL:PC) Nanodisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
