3AA0
 
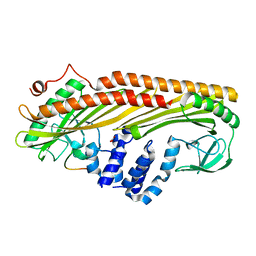 | | Crystal structure of Actin Capping Protein in complex with the Cp-binding motif derived from CARMIL | | Descriptor: | 21mer peptide from Leucine-rich repeat-containing protein 16A, CARBONATE ION, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
3AA1
 
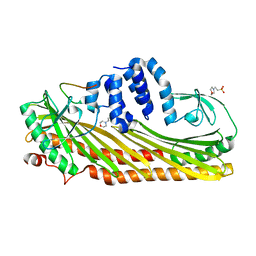 | | Crystal structure of Actin capping protein in complex with the Cp-binding motif derived from CKIP-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 23mer peptide from Pleckstrin homology domain-containing family O member 1, F-actin-capping protein subunit alpha-1, ... | | Authors: | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
2DW2
 
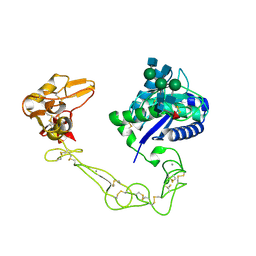 | | Crystal structure of VAP2 from Crotalus atrox venom (Form 2-5 crystal) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Catrocollastatin, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2006-08-02 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Febs Lett., 581, 2007
|
|
2DW0
 
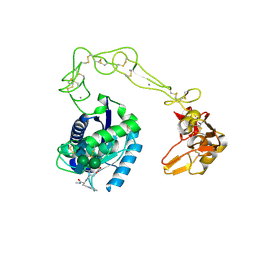 | | Crystal structure of VAP2 from Crotalus atrox venom (Form 2-1 crystal) | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, CALCIUM ION, Catrocollastatin, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2006-08-02 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Febs Lett., 581, 2007
|
|
2DW1
 
 | | Crystal structure of VAP2 from Crotalus atrox venom (Form 2-2 crystal) | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, CALCIUM ION, Catrocollastatin, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2006-08-02 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Febs Lett., 581, 2007
|
|
3AQJ
 
 | | Crystal Structure of a C-terminal domain of the bacteriophage P2 tail spike protein, gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Takeda, S, Yamashita, E, Nakagawa, A. | | Deposit date: | 2010-11-06 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2E3X
 
 | | Crystal structure of Russell's viper venom metalloproteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, ... | | Authors: | Igarashi, T, Takeda, S. | | Deposit date: | 2006-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases.
Febs Lett., 581, 2007
|
|
7W51
 
 | |
7W4Z
 
 | |
7W50
 
 | |
7W52
 
 | |
7YNE
 
 | |
5YDN
 
 | | Mu pahge neck subunit | | Descriptor: | Gene product J | | Authors: | Takeda, S, Iwasaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-09-13 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structures of bacteriophage neck subunits are shared in Podoviridae, Siphoviridae and Myoviridae
Genes Cells, 23, 2018
|
|
5YVQ
 
 | | Complex of Mu phage tail fiber and its chaperone | | Descriptor: | GLYCEROL, Tail fiber assembly protein U, Tail fiber protein S | | Authors: | Takeda, S, Sakai, K, Iwazaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-11-27 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
Nat Microbiol, 4, 2019
|
|
2BEC
 
 | | Crystal structure of CHP2 in complex with its binding region in NHE1 and insights into the mechanism of pH regulation | | Descriptor: | Calcineurin B homologous protein 2, Sodium/hydrogen exchanger 1, YTTRIUM (III) ION | | Authors: | Ben Ammar, Y, Takeda, S, Hisamitsu, T, Mori, H, Wakabayashi, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation
Embo J., 25, 2006
|
|
3GHM
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
3GHN
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
3SBK
 
 | | Russell's viper venom serine proteinase, RVV-V (PPACK-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Vipera russelli proteinase RVV-V gamma | | Authors: | Nakayama, D, Ben Ammar, Y, Takeda, S. | | Deposit date: | 2011-06-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of coagulation factor V recognition for cleavage by RVV-V
Febs Lett., 585, 2011
|
|
1A2X
 
 | | COMPLEX OF TROPONIN C WITH A 47 RESIDUE (1-47) FRAGMENT OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, TROPONIN I | | Authors: | Vassylyev, D.G, Takeda, S, Wakatsuki, S, Maeda, K, Maeda, Y. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8JU3
 
 | | Mu phage tail fiber | | Descriptor: | Tail fiber protein S,Tail fiber protein S' | | Authors: | Yamashita, E, Takeda, S. | | Deposit date: | 2023-06-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of the three-dimensional structure of bacteriophage Mu(-) tail fiber and its characterization.
Virology, 593, 2024
|
|
2D4C
 
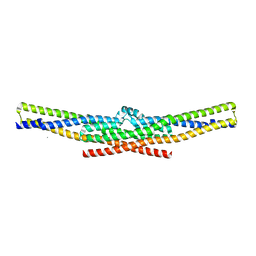 | | Crystal structure of the endophilin BAR domain mutant | | Descriptor: | CALCIUM ION, SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S. | | Deposit date: | 2005-10-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
3ABE
 
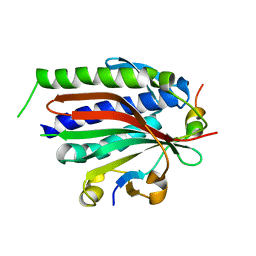 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
1ET0
 
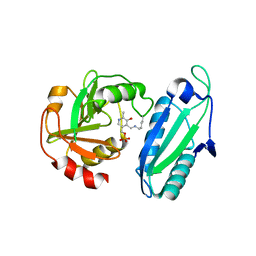 | | CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Nakai, T, Mizutani, H, Miyahara, I, Hirotsu, K, Takeda, S, Jhee, K.H, Yoshimura, T, Esaki, N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli.
J.Biochem.(Tokyo), 128, 2000
|
|
8GSU
 
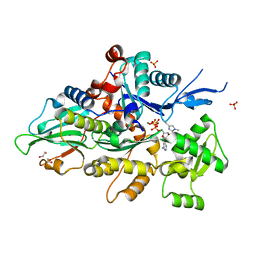 | | Crystal structure of human cardiac alpha actin (WT_ADP-Pi) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT4
 
 | | Crystal structure of human cardiac alpha actin Q137A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
