2MW3
 
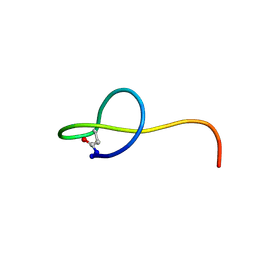 | | Solution NMR structure of the lasso peptide streptomonomicin | | Descriptor: | Lasso peptide | | Authors: | Tietz, J.I, Zhu, L, Mitchell, D.A, Metelev, M, Melby, J.O, Blair, P.M, Livnat, I, Severinov, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso Peptide from an understudied halophilic actinomycete.
Chem.Biol., 22, 2015
|
|
5K99
 
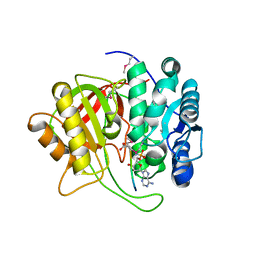 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
5NJA
 
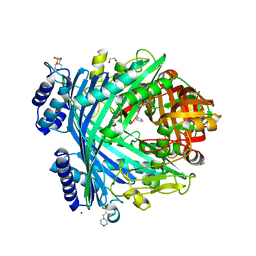 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJF
 
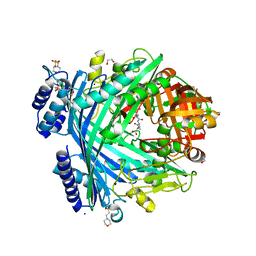 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJB
 
 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJC
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD E263A mutant) with hexapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metalloprotease PmbA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ5
 
 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJ9
 
 | | E. coli Microcin-processing metalloprotease TldD/E with DRVY angiotensin fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ASP-ARG-VAL-TYR, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
6IBG
 
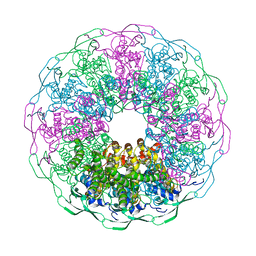 | | Bacteriophage G20c portal protein crystal structure for construct with intact N-terminus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Portal protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6I9E
 
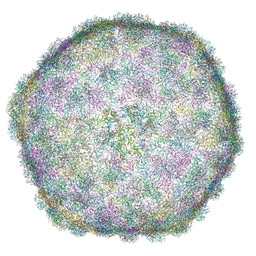 | | Thermophage P23-45 empty expanded capsid | | Descriptor: | Auxiliary protein, Major head protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6IBC
 
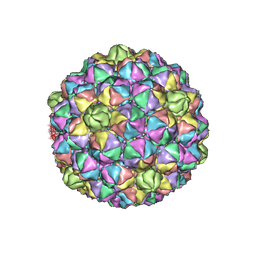 | | Thermophage P23-45 procapsid | | Descriptor: | Major head protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3WOF
 
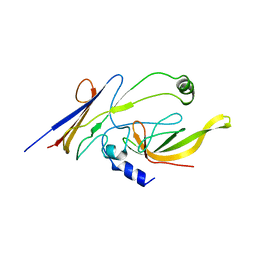 | | Crystal structure of P23-45 gp39 (6-132) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
3WOE
 
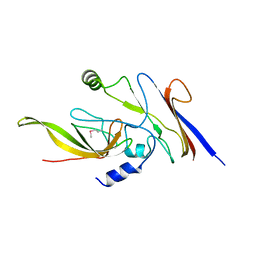 | | Crystal structure of P23-45 gp39 (6-109) bound to Thermus thermophilus RNA polymerase beta-flap domain | | Descriptor: | DNA-directed RNA polymerase subunit beta, Putative uncharacterized protein | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
3WOD
 
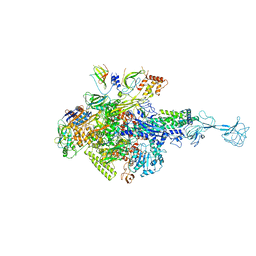 | | RNA polymerase-gp39 complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
1SIG
 
 | |
3GJZ
 
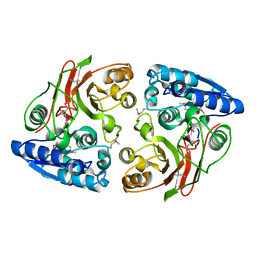 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis str. Ames | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-09 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
5LGM
 
 | | Gp5.7 mutant L42A | | Descriptor: | Fusion protein 5.5/5.7 | | Authors: | Liu, B, Matthews, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gp5.7 mutant L42A
to be published
|
|
2GHO
 
 | | Recombinant Thermus aquaticus RNA polymerase for Structural Studies | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta' | | Authors: | Lamour, V, Darst, S.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Recombinant Thermus aquaticus RNA Polymerase for Structural Studies.
J.Mol.Biol., 359, 2006
|
|
6QLC
 
 | | The ssDNA-binding RNA polymerase cofactor Drc from Pseudomonas phage LUZ7 | | Descriptor: | PHOSPHATE ION, ssDNA binding RNA Polymerase cofactor | | Authors: | De Zitter, E, Boon, M, De Smet, J, Lavigne, R, Van Meervelt, L. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 'Drc', a structurally novel ssDNA-binding transcription regulator of N4-related bacterial viruses.
Nucleic Acids Res., 48, 2020
|
|
8BB1
 
 | |
3BMB
 
 | |
8UX9
 
 | | Asymmetric unit of the PARIS Immune Complex at 3.2 Angstrom Resolution | | Descriptor: | AriA, AriB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Burman, N.B, Henriques, W, Wilkinson, R, Graham, A, Wiedenheft, B. | | Deposit date: | 2023-11-09 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A virally encoded tRNA neutralizes the PARIS antiviral defence system.
Nature, 634, 2024
|
|
7Z6F
 
 | |
7Z8E
 
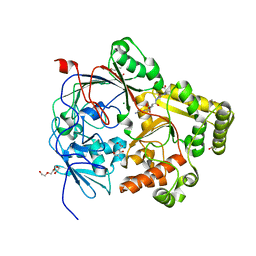 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | Authors: | Morera, S, Vigouroux, V, Travin, D.Y. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
6OM4
 
 | |
