3ZX4
 
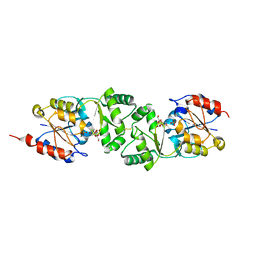 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate,orthophosphate and magnesium | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZX5
 
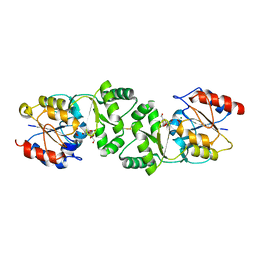 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, covalently bound to vanadate and in complex with alpha- mannosylglycerate and magnesium | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
4V2X
 
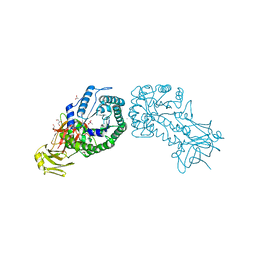 | | High resolution structure of the full length tri-modular endo-beta-1, 4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4UZ8
 
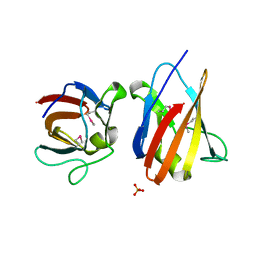 | | The SeMet structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B), SULFATE ION | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
6WM5
 
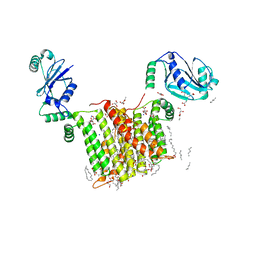 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-04-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
2LKV
 
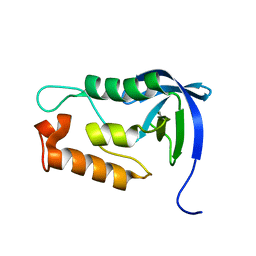 | | Staphylococcal Nuclease PHS variant | | Descriptor: | Thermonuclease | | Authors: | Matzapetakis, M, Pais, T.M, Lamosa, P, Turner, D.L, Santos, H. | | Deposit date: | 2011-10-21 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mannosylglycerate stabilizes staphylococcal nuclease with restriction of slow beta-sheet motions.
Protein Sci., 21, 2012
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
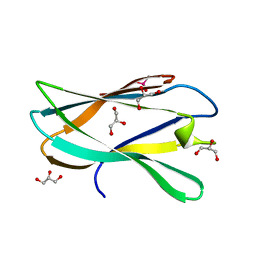 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6WMV
 
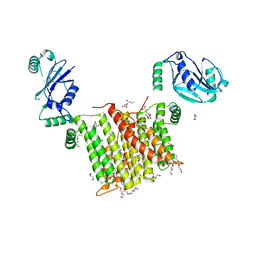 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,3',3''-phosphanetriyltripropanoic acid, AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F. | | Deposit date: | 2020-04-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
1E5D
 
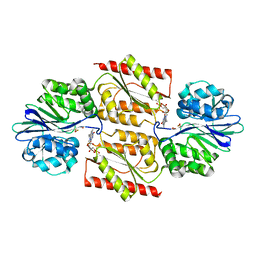 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
1QN1
 
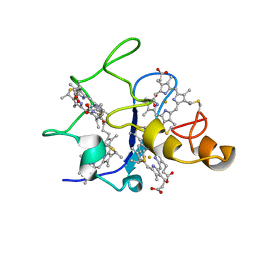 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
2BPN
 
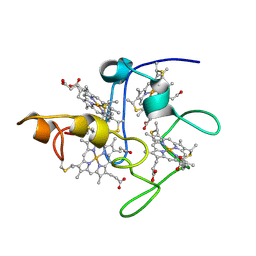 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERRICYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Aguiar, A.P, Brennan, L, Xavier, A.V, Turner, D.L. | | Deposit date: | 2005-04-21 | | Release date: | 2006-03-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Tetrahaem Ferricytochrome C(3) from Desulfovibrio Vulgaris (Hildenborough) and its K45Q Mutant: The Molecular Basis of Cooperativity.
Biochim.Biophys.Acta, 1757, 2006
|
|
