6CG9
 
 | | Crystal structure of Triosephosphate Isomerase from Zea mays (mexican corn) | | Descriptor: | ACETATE ION, GLYCEROL, Triosephosphate isomerase, ... | | Authors: | Romero-Romero, S, Fernandez-Velasco, D.A, Rodriguez-Romero, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and conformational stability of the triosephosphate isomerase from Zea mays. Comparison with the chemical unfolding pathways of other eukaryotic TIMs.
Arch. Biochem. Biophys., 658, 2018
|
|
7KOT
 
 | |
7KP5
 
 | |
1CI1
 
 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8EYM
 
 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM SHEWANELLA DENITRIFICANS OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AND N-ACETYLGLUCOSAMINE-6-PHOSPHATE AT 2.31 A RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rodriguez-Hernandez, A, Marcos-Viquez, J, Rodriguez-Romero, A, Bustos-Jaimes, I. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
8EOL
 
 | |
6XKS
 
 | | Crystal structure of domain A from the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) of Salmonella typhimurium | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Histidine ABC transporter substrate-binding protein HisJ | | Authors: | Romero-Romero, S, Berrocal, T, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and kinetic analysis of the LAO binding protein and its isolated domains reveal non-additivity in stability, folding and function.
Febs J., 2023
|
|
5FEF
 
 | |
5FEG
 
 | |
5FDS
 
 | |
4GNY
 
 | | Bovine beta-lactoglobulin complex with dodecyl sulfate | | Descriptor: | Beta-lactoglobulin, DODECYL SULFATE, GLYCEROL | | Authors: | Gutierrez-Magdaleno, G, Torres-Rivera, A, Garcia-Hernandez, E, Rodriguez-Romero, A. | | Deposit date: | 2012-08-17 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6367 Å) | | Cite: | Ligand-binding and self-association cooperativity of beta-lactoglobulin
J.Mol.Recognit., 26, 2013
|
|
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6UTM
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6UL3
 
 | |
6UKC
 
 | |
4Y9A
 
 | |
4Y8F
 
 | |
4Y96
 
 | | Crystal structure of Triosephosphate Isomerase from Gemmata obscuriglobus | | Descriptor: | CALCIUM ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
4Y90
 
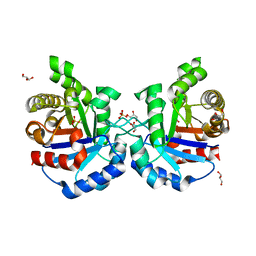 | | Crystal structure of Triosephosphate Isomerase from Deinococcus radiodurans | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Romero-Romero, S, Rodriguez-Romero, A, Fernadez-Velasco, D.A. | | Deposit date: | 2015-02-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Phys Chem Chem Phys, 17, 2015
|
|
8V2X
 
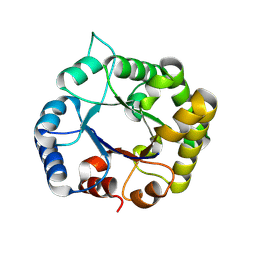 | |
8V0A
 
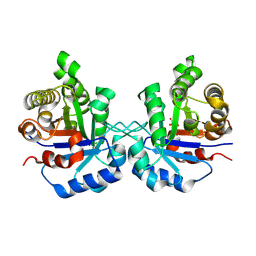 | | Crystal Structure of the worst case of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V09
 
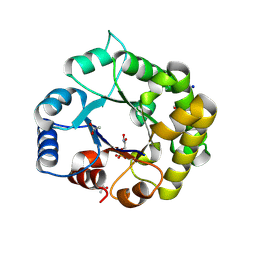 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by bayesian inference with PGH | | Descriptor: | ACETIC ACID, FORMIC ACID, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8V2W
 
 | | Crystal Structure of the ancestral triosephosphate isomerase reconstruction of the last opisthokont common ancestor obtained by Bayesian inference | | Descriptor: | GLYCEROL, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
8W08
 
 | | Crystal Structure of the worst case reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood | | Descriptor: | FLUORIDE ION, Triosephosphate isomerase | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
