1G4K
 
 | | X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin | | Descriptor: | 5-METHYL-5-(4-PHENOXY-PHENYL)-PYRIMIDINE-2,4,6-TRIONE, CALCIUM ION, GLYCEROL, ... | | Authors: | Dunten, P, Kammlott, U, Crowther, R, Levin, W, Foley, L.H, Wang, P, Palermo, R. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin.
Protein Sci., 10, 2001
|
|
8CGM
 
 | | Structure of the lipoprotein transporter LolA from Porphyromonas gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Persson, K, Jaiman, D, Nagampalli, R. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A comparative analysis of lipoprotein transport proteins: LolA and LolB from Vibrio cholerae and LolA from Porphyromonas gingivalis.
Sci Rep, 13, 2023
|
|
8CHX
 
 | |
5DZQ
 
 | | C3larvin toxin, an ADP-ribosyltransferase from Paenibacillus larvae, Orthorhombic Form | | Descriptor: | GLYCEROL, MAGNESIUM ION, Toxin-like protein | | Authors: | Dutta, D, Ravulapalli, R, Merrill, A.R. | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural variability of C3larvin toxin. Intrinsic dynamics of the alpha / beta fold of the C3-like group of mono-ADP-ribosyltransferase toxins.
J. Biomol. Struct. Dyn., 34, 2016
|
|
2MMZ
 
 | | Solution structure of the apo form of human glutaredoxin 5 | | Descriptor: | Glutaredoxin-related protein 5, mitochondrial | | Authors: | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9BC9
 
 | | Structure of KLHDC2 bound to SJ10278 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[2-chloro-4-(methoxymethoxy)phenyl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
6TAO
 
 | | The cytotoxin MakE from Vibrio cholerae | | Descriptor: | NICKEL (II) ION, Non-hemolytic enterotoxin lytic component L1, SULFATE ION | | Authors: | Persson, K, Nagampalli, R, Heidler, T, Wai, S.N. | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A tripartite cytolytic toxin formed by Vibrio cholerae proteins with flagellum-facilitated secretion.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6TCT
 
 | |
9BCA
 
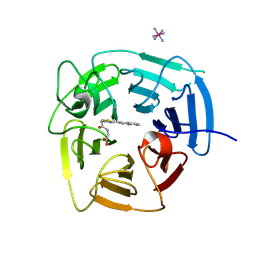 | | Structure of KLHDC2 bound to SJ46411 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-{[2-(naphthalen-2-yl)-1,3-thiazol-4-yl]acetyl}glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
9BCC
 
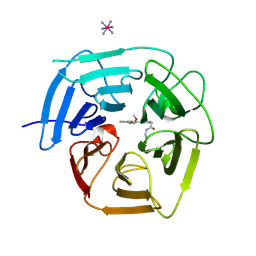 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
6T8D
 
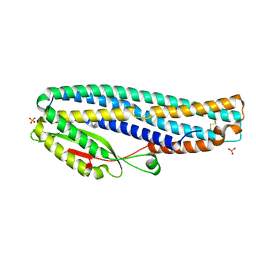 | | The cytotoxin MakB from Vibrio cholerae | | Descriptor: | MakB, SULFATE ION | | Authors: | Persson, K, Nagampalli, R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A tripartite cytolytic toxin formed by Vibrio cholerae proteins with flagellum-facilitated secretion.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1ZK6
 
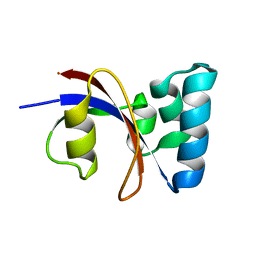 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
2QFE
 
 | | Distal C2-Like Domain of Human Calpain-7 | | Descriptor: | BROMIDE ION, Calpain-7 | | Authors: | Walker, J.R, Cuerrier, D, Ravulapalli, R, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the C2 Domain of Human Calpain-7
To be Published
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFI
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFM
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4OKH
 
 | | Crystal structure of calpain-3 penta-EF-hand domain | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CALCIUM ION, Calpain-3 | | Authors: | Karunan Partha, S, Ravulapalli, R, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of calpain-3 penta-EF-hand (PEF) domain - a homodimerized PEF family member with calcium bound at the fifth EF-hand.
Febs J., 281, 2014
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
2I7A
 
 | | Domain IV of Human Calpain 13 | | Descriptor: | CALCIUM ION, Calpain 13, GLYCEROL, ... | | Authors: | Walker, J.R, Ng, K, Davis, T.L, Ravulapalli, R, Butler-cole, C, Finerty Jr, P.J, Newman, E.M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Calpain 13
To be Published
|
|
1RPG
 
 | | STRUCTURES OF RNASE A COMPLEXED WITH 3'-CMP AND D(CPA): ACTIVE SITE CONFORMATION AND CONSERVED WATER MOLECULES | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, RIBONUCLEASE A | | Authors: | Zegers, I, Wyns, L, Palmer, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structures of RNase A complexed with 3'-CMP and d(CpA): active site conformation and conserved water molecules.
Protein Sci., 3, 1994
|
|
1RPF
 
 | |
1RPH
 
 | |
3RHK
 
 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
3RHX
 
 | | Crystal structure of the catalytic domain of FGFR1 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, 1,2-ETHANEDIOL, Basic fibroblast growth factor receptor 1, ... | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
