8HYH
 
 | | Structure of amino acid dehydrogenase3448 | | Descriptor: | 1,2-ETHANEDIOL, Alanine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
6K9Z
 
 | | STRUCTURE OF URIDYLYLTRANSFERASE MUTANT | | Descriptor: | ACETATE ION, FE (III) ION, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
2D4A
 
 | | Structure of the malate dehydrogenase from Aeropyrum pernix | | Descriptor: | Malate dehydrogenase | | Authors: | Kawakami, R, Sakuraba, H, Tsuge, H, Ohshima, T. | | Deposit date: | 2005-10-12 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Refolding, characterization and crystal structure of (S)-malate dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
Biochim.Biophys.Acta, 1794, 2009
|
|
6JXN
 
 | | Crystal Structure of Indigo reductase from Bacillus smithii type strain DSM 4216 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and biochemical characterization of an extremely thermostable FMN-dependent NADH-indigo reductase from Bacillus smithii.
Int.J.Biol.Macromol., 164, 2020
|
|
6JXS
 
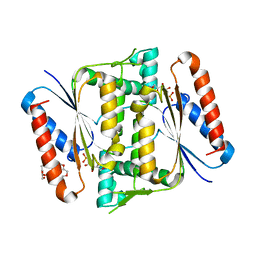 | | Crystal Structure of Indigo reductase (Y151F) from Bacillus smithii type strain DSM 4216 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical characterization of an extremely thermostable FMN-dependent NADH-indigo reductase from Bacillus smithii.
Int.J.Biol.Macromol., 164, 2020
|
|
3AW5
 
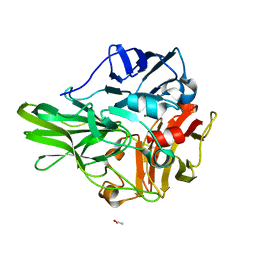 | | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum
Acta Crystallogr.,Sect.F, 67, 2011
|
|
8J1G
 
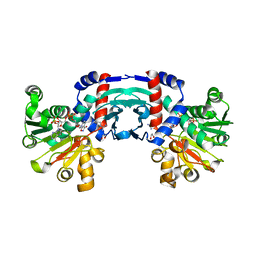 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
8J1C
 
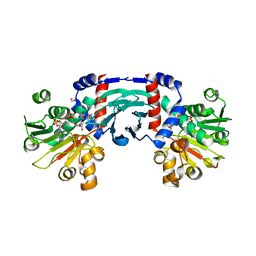 | |
5WYF
 
 | | Structure of amino acid racemase, 2.12 A | | Descriptor: | CADMIUM ION, Isoleucine 2-epimerase, N-[O-PHOSPHONO-PYRIDOXYL]-ISOLEUCINE | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1V9L
 
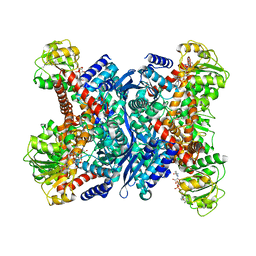 | | L-glutamate dehydrogenase from Pyrobaculum islandicum complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glutamate dehydrogenase | | Authors: | Bhuiya, M.W, Sakuraba, H, Ohshima, T, Imagawa, T, Katunuma, N, Tsuge, H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The First Crystal Structure of Hyperthermostable NAD-dependent Glutamate Dehydrogenase from Pyrobaculum islandicum
J.Mol.Biol., 345, 2005
|
|
5WYA
 
 | | Structure of amino acid racemase, 2.65 A | | Descriptor: | (2S,3S)-3-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pentanoic acid, DIMETHYL SULFOXIDE, Isoleucine 2-epimerase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3ABI
 
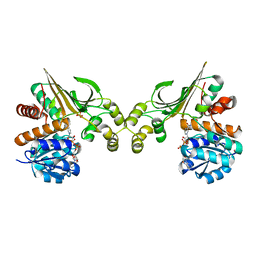 | | Crystal Structure of L-Lysine Dehydrogenase from Hyperthermophilic Archaeon Pyrococcus horikoshii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative uncharacterized protein PH1688, SULFATE ION | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, J, Ohshima, T. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | First crystal structure of L-lysine 6-dehydrogenase as an NAD-dependent amine dehydrogenase.
J.Biol.Chem., 285, 2010
|
|
1L2L
 
 | |
3W6U
 
 | |
3WS7
 
 | | The 1.18 A resolution structure of L-serine 3-dehydrogenase complexed with NADP+ and sulfate ion from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | 6-phosphogluconate dehydrogenase, NAD-binding protein, ACETIC ACID, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2014-03-04 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the NADP+and tartrate-bound complex of L-serine 3-dehydrogenase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Extremophiles, 22, 2018
|
|
3WYC
 
 | | Structure of a meso-diaminopimelate dehydrogenase in complex with NADP | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WYB
 
 | | Structure of a meso-diaminopimelate dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FB3
 
 | | Structure of glycerophosphate dehydrogenase in complex with NADPH | | Descriptor: | Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE, ... | | Authors: | Sakuraba, H, Hayashi, J, Yamamoto, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique coenzyme binding mode of hyperthermophilic archaeal sn-glycerol-1-phosphate dehydrogenase from Pyrobaculum calidifontis
Proteins, 84, 2016
|
|
5B1Y
 
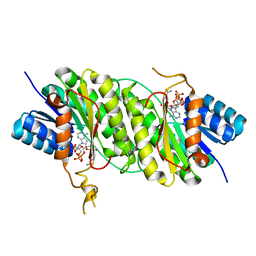 | | Crystal structure of NADPH bound carbonyl reductase from Aeropyrum pernix | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Araki, T, Ohshima, T. | | Deposit date: | 2015-12-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic properties and crystal structure of thermostable NAD(P)H-dependent carbonyl reductase from the hyperthermophilic archaeon Aeropyrum pernix K1.
Enzyme.Microb.Technol., 91, 2016
|
|
4YSV
 
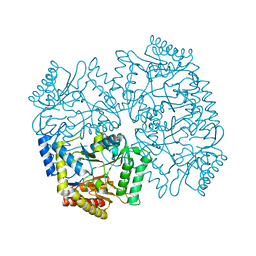 | |
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8HMO
 
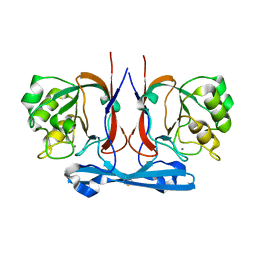 | |
3A9W
 
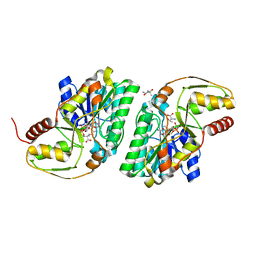 | | Crystal structure of L-Threonine bound L-Threonine dehydrogenase (Y137F) from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
3A4V
 
 | | Crystal structure of pyruvate bound L-Threonine dehydrogenase from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
2YY7
 
 | | Crystal structure of thermolabile L-threonine dehydrogenase from Flavobacterium frigidimaris KUC-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, ... | | Authors: | Yoneda, K, Sakuraba, H, Oikawa, T, Muraoka, I, Ohshima, T. | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Crystal structure of UDP-galactose 4-epimerase-like L-threonine dehydrogenase belonging to the intermediate short-chain dehydrogenase-reductase superfamily
Febs J., 277, 2010
|
|
