1KO7
 
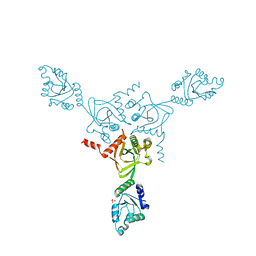 | | X-ray structure of the HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution | | Descriptor: | Hpr kinase/phosphatase, PHOSPHATE ION | | Authors: | Marquez, J.A, Hasenbein, S, Koch, B, Fieulaine, S, Nessler, S, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5JUF
 
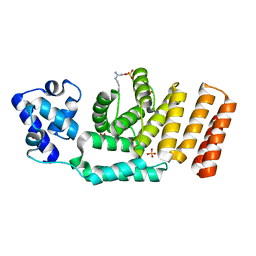 | | Crystal structure of the apo form of ComR from S. thermophilus. | | Descriptor: | SULFATE ION, Transcriptional regulator | | Authors: | Talagas, A, Fontaine, L, Ledesma, L, Li de la Sierra-Gallay, I, Hols, P, Nessler, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
PLoS Pathog., 12, 2016
|
|
5JUB
 
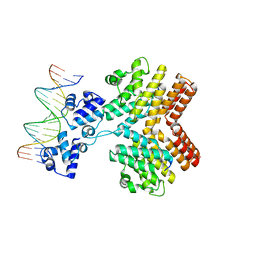 | | Crystal structure of ComR from S.thermophilus in complex with DNA and its signalling peptide ComS. | | Descriptor: | ComS, Transcriptional regulator, pComX-for, ... | | Authors: | Talagas, A, Fontaine, L, Ledesma-Garcia, L, Li de la Sierra-Gallay, I, Hols, P, Nessler, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
PLoS Pathog., 12, 2016
|
|
4FSC
 
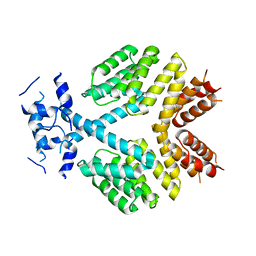 | | Crystal Structure of Bacillus thuringiensis PlcR in its apo form | | Descriptor: | Transcriptional activator PlcR protein | | Authors: | Grenha, R, Slamti, L, Bouillaut, L, Lereclus, D, Nessler, S. | | Deposit date: | 2012-06-27 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2FEP
 
 | | Structure of truncated CcpA in complex with P-Ser-HPr and Sulfate ions | | Descriptor: | Catabolite control protein A, Phosphocarrier protein HPr, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Poncet, S, Lecampion, C, Meyer, P, Deutscher, J, Galinier, A, Nessler, S. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of B. subtilis CcpA effector binding site.
Proteins, 64, 2006
|
|
7ZPT
 
 | |
7ZPU
 
 | |
8AZG
 
 | |
8AAM
 
 | |
8AB4
 
 | |
5FD4
 
 | |
2GA8
 
 | |
2GAA
 
 | |
