8QND
 
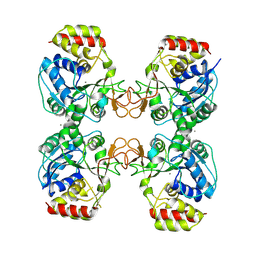 | | Crystal structure of the ribonucleoside hydrolase C from Lactobacillus reuteri | | Descriptor: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase | | Authors: | Matyuta, I.O, Minyaev, M.E, Shaposhnikov, L.A, Pometun, A.A, Tishkov, V.I, Popov, V.O, Boyko, K.M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Functional Examination of Novel Ribonucleoside Hydrolase C (RihC) from Limosilactobacillus reuteri LR1.
Int J Mol Sci, 25, 2023
|
|
8RAF
 
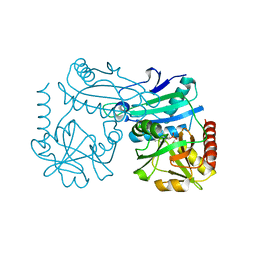 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8RAI
 
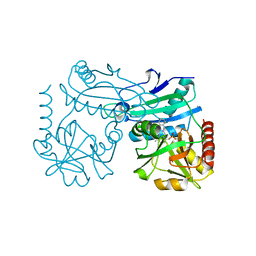 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
8QPT
 
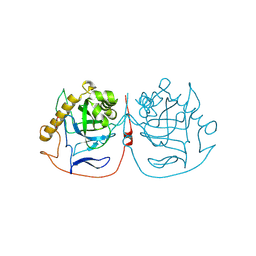 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
9UJ2
 
 | | 14-3-3 zeta chimera with the S202R peptide of SARS-CoV-2 N (residues 200-213) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta,Peptide from Nucleoprotein, DI(HYDROXYETHYL)ETHER | | Authors: | Boyko, K.M, Matyuta, I.O, Minyaev, M.E, Perfilova, K.V, Sluchanko, N.N. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Biochem.Biophys.Res.Commun., 767, 2025
|
|
7Z79
 
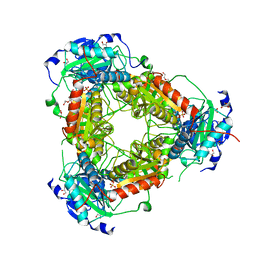 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
8ZBO
 
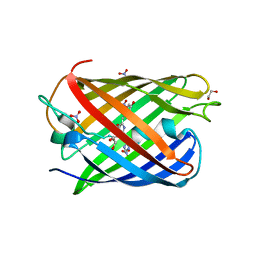 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
9J32
 
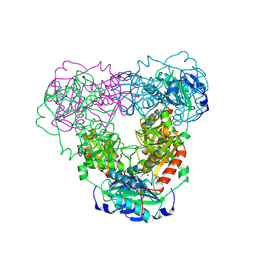 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus mutant N174K | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER | | Authors: | Ilyasov, I.O, Minyaev, M.E, Rakitina, T.V, Bakunova, A.K, Matyuta, I.O, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein of unknown function from variovorax paradoxus with amino acid substitution n174k is able to form schiff base with plp molecule
Kristallografiya, 69, 2024
|
|
8PEI
 
 | | Crystal structure of the biphotochromic fluorescent protein SAASoti (C21N/V127T variant) in its green on-state | | Descriptor: | C21N/V127T form of the biphotochromic fluorescent protein SAASoti | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Matyuta, I.O, Gavshina, A.V, Solovyev, I.D, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of the correlated motion(s) of the chromophore in photoswitching of green and red forms of the photoconvertible fluorescent protein mSAASoti.
Sci Rep, 14, 2024
|
|
