7RMC
 
 | | Yeast CTP Synthase (Ura7) filament bound to CTP at low pH | | Descriptor: | CTP synthase 1, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNR
 
 | | Yeast CTP Synthase (Ura8) Bundle Bound to Substrates at Low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RL5
 
 | | Yeast CTP Synthase (URA8) filament bound to CTP at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RMO
 
 | | Yeast CTP Synthase (Ura7) Bundle bound to Products at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RL0
 
 | | Yeast CTP Synthase (URA8) Filament bound to ATP/UTP at low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7RNL
 
 | | Yeast CTP Synthase (Ura7) H360R Filament bound to Substrates | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
6E9V
 
 | | DHF79 filament | | Descriptor: | DHF79 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9Y
 
 | | DHF38 filament | | Descriptor: | DHF38 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9T
 
 | | DHF58 filament | | Descriptor: | DHF58 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9Z
 
 | | DHF119 filament | | Descriptor: | DHF119 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9R
 
 | | DHF46 filament | | Descriptor: | DHF46 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
5TKV
 
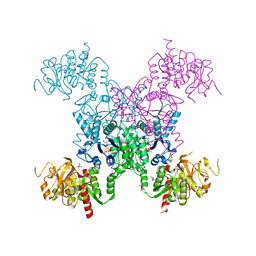 | | X-RAY CRYSTAL STRUCTURE OF THE "CLOSED" CONFORMATION OF CTP-INHIBITED E. COLI CYTIDINE TRIPHOSPHATE (CTP) SYNTHETASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, ... | | Authors: | Baldwin, E.P, Endrizzi, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6XQW
 
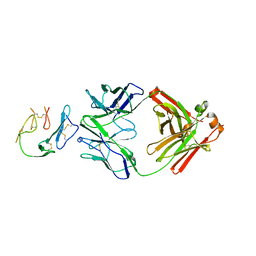 | | Crystal Structure of MaliM03 Fab in complex with Pfmsp1-19 | | Descriptor: | MaliM03 Fab Heavy Chain, MaliM03 Fab Light Chain, Pfmsp1-19 | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites.
J.Exp.Med., 218, 2021
|
|
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6TJ1
 
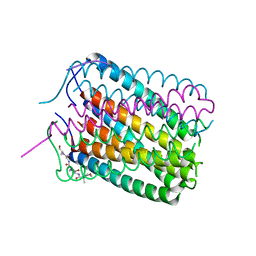 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | De novo designed WSHC6, purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-11-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6P4X
 
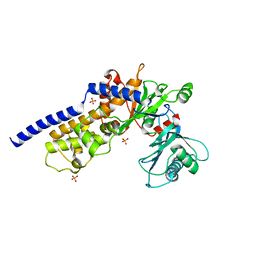 | |
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6O35
 
 | |
