2MEO
 
 | |
5L57
 
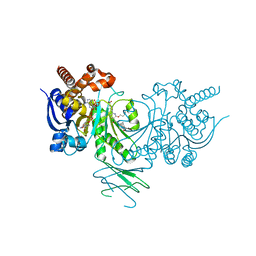 | | Crystal structure of Iso-citrate Dehydrogenase R132H in complex with a novel inhibitor (compound 13a) | | Descriptor: | (1~{R},5~{S})-3-[6-(3-methylbutoxy)-5-[[(1~{R},3~{S})-5-oxidanyl-2-adamantyl]carbamoyl]pyridin-2-yl]-3-azabicyclo[3.1.0]hexane-6-carboxylic acid, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Levy, C. | | Deposit date: | 2016-05-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Discovery and Optimization of Allosteric Inhibitors of Mutant Isocitrate Dehydrogenase 1 (R132H IDH1) Displaying Activity in Human Acute Myeloid Leukemia Cells.
J.Med.Chem., 59, 2016
|
|
5L58
 
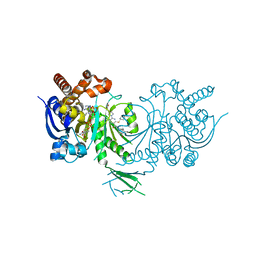 | | Crystal structure of Iso-citrate Dehydrogenase 1 [IDH1 (R132H)] in complex with a novel inhibitor (Compound 2) | | Descriptor: | 2-[(3~{R})-1-[6-cyclohexylsulfanyl-5-[[(1~{R},3~{S})-5-oxidanyl-2-adamantyl]carbamoyl]pyridin-2-yl]pyrrolidin-3-yl]ethanoic acid, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Levy, C. | | Deposit date: | 2016-05-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Discovery and Optimization of Allosteric Inhibitors of Mutant Isocitrate Dehydrogenase 1 (R132H IDH1) Displaying Activity in Human Acute Myeloid Leukemia Cells.
J.Med.Chem., 59, 2016
|
|
7ZPT
 
 | |
7ZPU
 
 | |
8AZG
 
 | |
2KY3
 
 | |
2LO7
 
 | |
2LU9
 
 | |
1WRA
 
 | |
2IEH
 
 | |
