3CMD
 
 | | Crystal structure of peptide deformylase from VRE-E.faecium | | Descriptor: | FE (III) ION, MALONATE ION, Peptide deformylase, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2008-03-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure.
Proteins, 74, 2009
|
|
3CMJ
 
 | |
3FAK
 
 | |
3E5P
 
 | | Crystal structure of alanine racemase from E.faecalis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine racemase, NONAETHYLENE GLYCOL, ... | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3DNM
 
 | |
3E6E
 
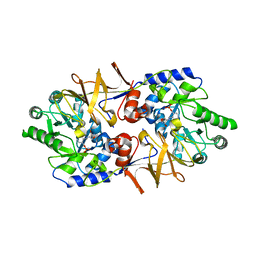 | | Crystal structure of Alanine racemase from E.faecalis complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
4K86
 
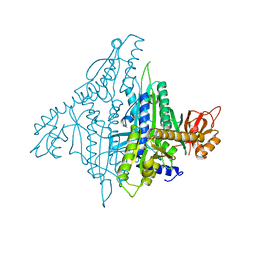 | |
4KP2
 
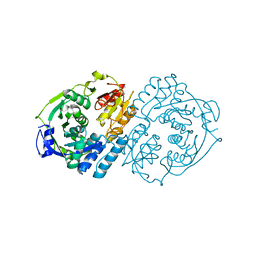 | |
4K87
 
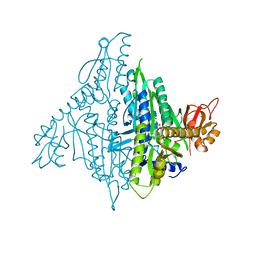 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LWL
 
 | |
4K88
 
 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KP1
 
 | | Crystal structure of IPM isomerase large subunit from methanococcus jannaschii (MJ0499) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,4-dimethylpentane-2,4-diol, Isopropylmalate/citramalate isomerase large subunit, ... | | Authors: | Hwang, K.Y, Lee, E.H. | | Deposit date: | 2013-05-13 | | Release date: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization and comparison of the large subunits of IPM isomerase and homoaconitase from Methanococcus jannaschii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LWM
 
 | |
4LWK
 
 | |
3H17
 
 | | Crystal structure of EstE5-PMSF (I) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3H18
 
 | | Crystal structure of EstE5-PMSF (II) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3Q4R
 
 | |
3Q4N
 
 | |
3II1
 
 | |
3Q4Q
 
 | |
3Q4O
 
 | |
3G6N
 
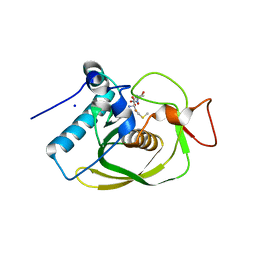 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2NX8
 
 | |
5HXY
 
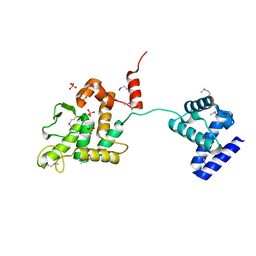 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
5HZT
 
 | | Crystal structure of Dronpa-Cu2+ | | Descriptor: | COPPER (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
