4DMZ
 
 | | PelD 156-455 from Pseudomonas aeruginosa PA14, apo form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
4F9J
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4WCJ
 
 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
2QSU
 
 | |
2C44
 
 | | Crystal Structure of E. coli Tryptophanase | | Descriptor: | POTASSIUM ION, SULFATE ION, TRYPTOPHANASE | | Authors: | Ku, S.-Y, Yip, P, Howell, P.L. | | Deposit date: | 2005-10-15 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia Coli Tryptophanase
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6OLJ
 
 | |
7SA8
 
 | |
6OK2
 
 | |
6OLL
 
 | |
7T8N
 
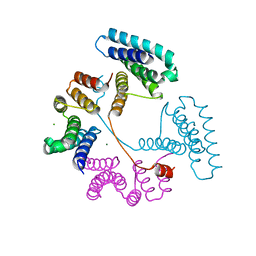 | | Crystal structure of the PNAG binding module PgaA-TPR 220-359 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Pfoh, R, Little, D.J, Howell, P.L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing.
Plos Pathog., 18, 2022
|
|
6OJZ
 
 | |
4DN0
 
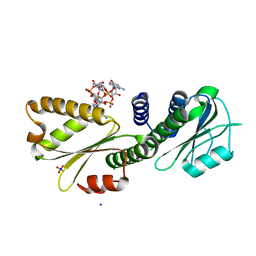 | | PelD 156-455 from Pseudomonas aeruginosa PA14 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein pelD, SODIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
6WJB
 
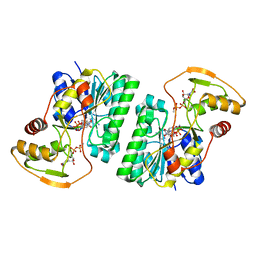 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
3O4V
 
 | |
3PG0
 
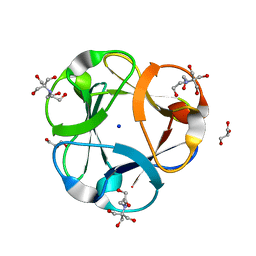 | | Crystal structure of designed 3-fold symmetric protein, ThreeFoil | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Lobsanov, Y.D, Broom, A, Howell, P.L, Rose, D.R, Meiering, E.M. | | Deposit date: | 2010-10-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein.
Structure, 20, 2012
|
|
3CQT
 
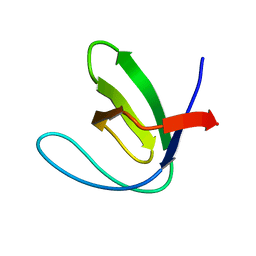 | | N53I V55L MUTANT of FYN SH3 DOMAIN | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Neculai, A.M, Zarrine-Afsar, A, Howell, P.L, Davidson, A, Chan, H.S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3DF9
 
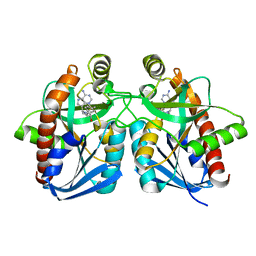 | |
3FZ2
 
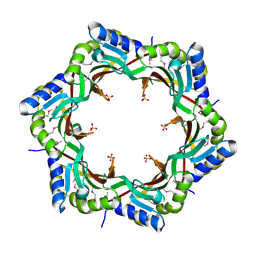 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
1KP3
 
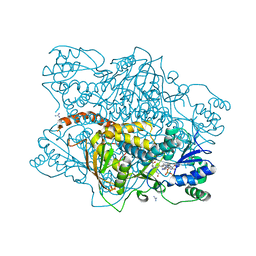 | |
1KP2
 
 | |
5TSY
 
 | |
5TCB
 
 | |
6AU1
 
 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2017-08-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
6BT4
 
 | | Crystal structure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit | | Descriptor: | 2-(acetylamino)-4-O-{2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranose, S-layer protein sap, SULFATE ION | | Authors: | Sychantha, D, Chapman, R.N, Bamford, N.C, Boons, G.J, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-12-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Molecular Basis for the Attachment of S-Layer Proteins to the Cell Wall of Bacillus anthracis.
Biochemistry, 57, 2018
|
|
6CZT
 
 | |
