9BIN
 
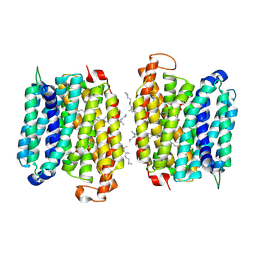 | | BRD-8000.3 bound EFPA transporter of Mycobacterium tuberculosis | | Descriptor: | (1S,3S)-N-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3-(2-methylprop-1-en-1-yl)cyclopropane-1-carboxamide, CHOLESTEROL HEMISUCCINATE, Uncharacterized MFS-type transporter EfpA, ... | | Authors: | Khandelwal, N.K, Gupta, M, Stroud, R.M. | | Deposit date: | 2024-04-23 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Inhibitor BRD-8000.3 bound EfpA transporter of Mycobacterium tuberculosis
To Be Published
|
|
9BII
 
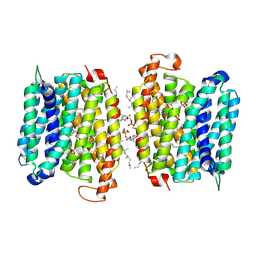 | | Mycobacterium tuberculosis EFPA antiparallel dimer | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Uncharacterized MFS-type transporter EfpA, [(2~{R})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadecanoyloxy-propyl] octadecanoate | | Authors: | Khandelwal, N.K, Gupta, M, Stroud, R.M. | | Deposit date: | 2024-04-23 | | Release date: | 2025-02-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mycobacterium tuberculosis EFPA antiparallel dimer
To Be Published
|
|
9BIQ
 
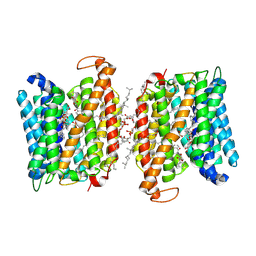 | | BRD-9327 bound EFPA transporter of Mycobacterium tuberculosis | | Descriptor: | CHOLESTEROL HEMISUCCINATE, N-(7-benzoyl-2,3-dihydro-1,4-benzodioxin-6-yl)-2-bromobenzamide, Uncharacterized MFS-type transporter EfpA, ... | | Authors: | Khandelwal, N.K, Gupta, M, Stroud, R.M. | | Deposit date: | 2024-04-24 | | Release date: | 2025-02-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | BRD-8000.3 bound EFPA transporter of Mycobacterium tuberculosis
To Be Published
|
|
7LER
 
 | | Netrin-1 filament assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Netrin-1, ... | | Authors: | McDougall, M, Gupta, M, Stetefeld, J. | | Deposit date: | 2021-01-14 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.99 Å) | | Cite: | The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation.
Nat Commun, 14, 2023
|
|
8TSY
 
 | | Pseudomonas fluorescens G150T-2 isocyanide hydratase at 274 K | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TT0
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TSZ
 
 | | Pseudomonas fluorescens G150T-3 isocyanide hydratase at 274 K | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TSU
 
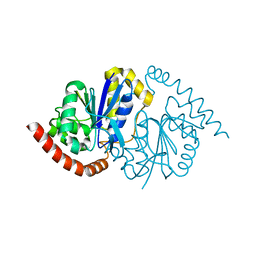 | | Pseudomonas fluorescens G150T-1 isocyanide hydratase at 274 K | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TSX
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 100 K | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8TT1
 
 | |
8F4F
 
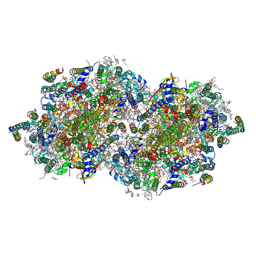 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4D
 
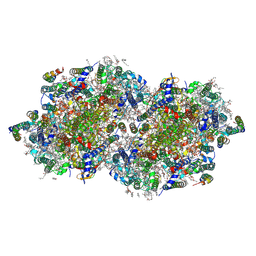 | | RT XFEL structure of Photosystem II 50 microseconds after the third illumination at 2.15 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8EZ5
 
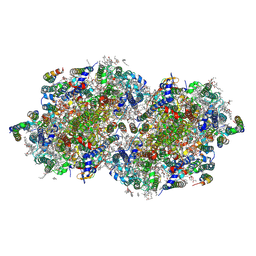 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4I
 
 | | RT XFEL structure of Photosystem II 2000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4H
 
 | | RT XFEL structure of Photosystem II 1200 microseconds after the third illumination at 2.10 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4C
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPW
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
