7PJ4
 
 | |
7PJ6
 
 | |
7PJ5
 
 | |
7PL2
 
 | |
7PL5
 
 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
8AIK
 
 | |
8AJI
 
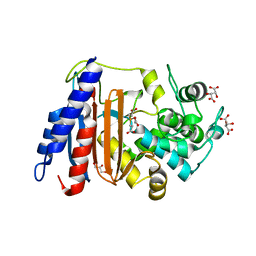 | | Crystal structure of DltE from L. plantarum, TCEP form | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Beta-lactamase family protein, GLYCEROL, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AKH
 
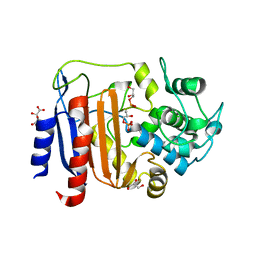 | | Crystal structure of DltE from L. plantarum soaked with LTA | | Descriptor: | Beta-lactamase family protein, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
8AGR
 
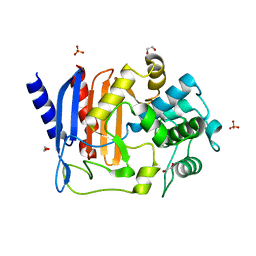 | | Crystal structure of DltE from L. plantarum, apo form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase family protein, SULFATE ION | | Authors: | Ravaud, S, Nikolopoulos, N, Grangeasse, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-function analysis of Lactiplantibacillus plantarum DltE& reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth.
Elife, 12, 2023
|
|
