4UCZ
 
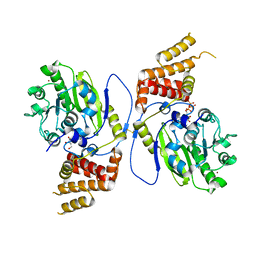 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | GUANOSINE, GUANOSINE-5'-TRIPHOSPHATE, RNA-DIRECTED RNA POLYMERASE L, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCJ
 
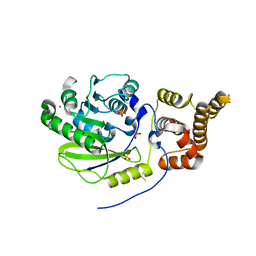 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCK
 
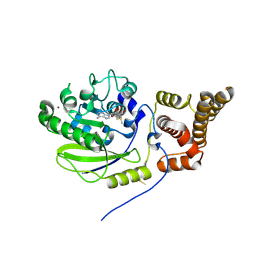 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCL
 
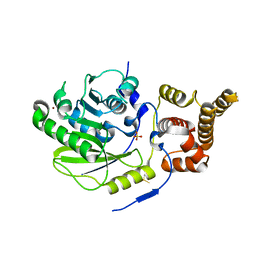 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, SULFATE ION, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
9FUX
 
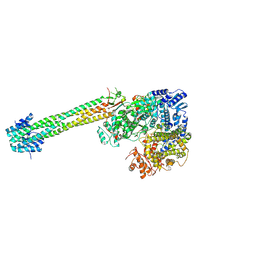 | | Cryo-EM structure of the Nipah virus polymerase (L) bound to the tetrameric phosphoprotein (P) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Balikci, E, Gunl, F, Carrique, L, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2024-06-26 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the Nipah virus polymerase complex.
Embo J., 44, 2025
|
|
9FTF
 
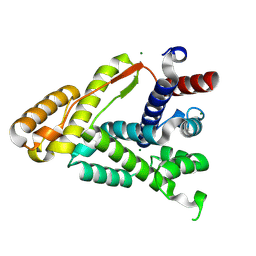 | | Crystal structure of the Connecting Domain of the Nipah virus Large protein | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Balikci, E, Gunl, F, Carrique, L, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2024-06-24 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Nipah virus polymerase complex.
Embo J., 44, 2025
|
|
4LP7
 
 | |
2CME
 
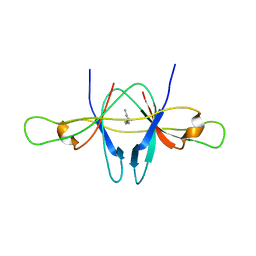 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
6I50
 
 | |
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
4XBU
 
 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | Descriptor: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XBR
 
 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | Authors: | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
5OIX
 
 | | Structure of the HMPV P oligomerization domain at 1.6 A | | Descriptor: | GLYCEROL, Phosphoprotein | | Authors: | Renner, M, Paesen, G.C, Grison, C.M, Granier, S, Grimes, J.M, Leyrat, C. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural dissection of human metapneumovirus phosphoprotein using small angle x-ray scattering.
Sci Rep, 7, 2017
|
|
5OIY
 
 | | Structure of the HMPV P oligomerization domain at 2.2 A | | Descriptor: | Phosphoprotein | | Authors: | Renner, M, Paesen, G.C, Grison, C.M, Granier, S, Grimes, J.M, Leyrat, C. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of human metapneumovirus phosphoprotein using small angle x-ray scattering.
Sci Rep, 7, 2017
|
|
8BCK
 
 | |
8BC5
 
 | |
8BCL
 
 | |
8BBT
 
 | |
6FLA
 
 | | 3H5 Fab bound to EDIII of DenV 2 Xtal form 1 | | Descriptor: | CHLORIDE ION, Domain III of Dengue virus 2, GLYCEROL, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
8BS7
 
 | |
6FLB
 
 | | 3H5 Fab bound to EDIII of DenV 2 Xtal form 2 | | Descriptor: | CHLORIDE ION, Domain III of Dengue virus 2, GLYCEROL, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
6FLC
 
 | | 2C8 Fab bound to EDIII of DenV 2 | | Descriptor: | Domain III of Dengue virus 2, GLYCEROL, Heavy chain of 2C8 Fab, ... | | Authors: | Flanagan, A, Renner, M, Grimes, J.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus.
Nat. Immunol., 19, 2018
|
|
3V6F
 
 | | Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unbound | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
3V6Z
 
 | | Crystal Structure of Hepatitis B Virus e-antigen | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain, e-antigen | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-20 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
4S1K
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 100 K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
