7SV6
 
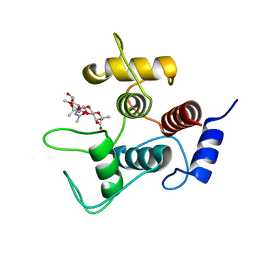 | |
7SV5
 
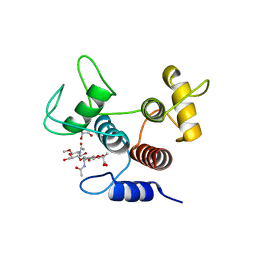 | | Crystal structure of SpaA-SLH/G109A in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
7SV3
 
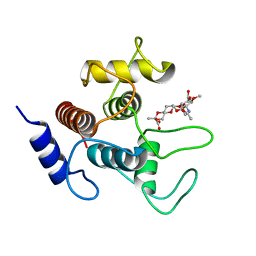 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
7SV4
 
 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAc-(1->3)-4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl-(1->4)-2-acetamido-2-deoxy-beta-D-glucopyranosyl-(1->3)-2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C9T
 
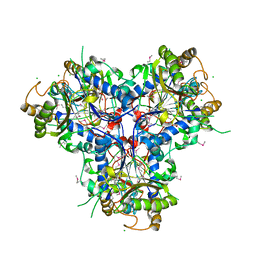 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
