6HH6
 
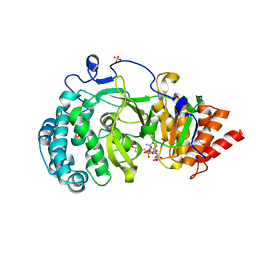 | | Human poly(ADP-ribose) glycohydrolase in complex with ADP-HPM | | Descriptor: | Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, Poly(ADP-ribose) glycohydrolase, SULFATE ION | | Authors: | Ariza, A. | | Deposit date: | 2018-08-24 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
5MKW
 
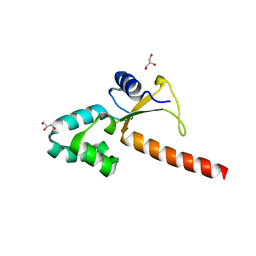 | | Crystal structure of the human ZRANB3 HNH domain | | Descriptor: | DNA annealing helicase and endonuclease ZRANB3, GLYCEROL, ZINC ION | | Authors: | Ariza, A. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the function of ZRANB3 in replication stress response.
Nat Commun, 8, 2017
|
|
5MLW
 
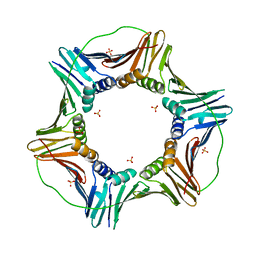 | |
5MLO
 
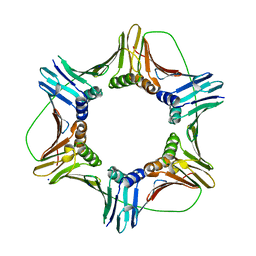 | |
5M31
 
 | | Macrodomain of Thermus aquaticus DarG | | Descriptor: | Appr-1-p processing domain protein, CHLORIDE ION, GLYCEROL | | Authors: | Ariza, A. | | Deposit date: | 2016-10-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
7OMU
 
 | | Thermosipho africanus DarTG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Macro domain-containing protein | | Authors: | Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
5M3I
 
 | | Macrodomain of Mycobacterium tuberculosis DarG | | Descriptor: | CHLORIDE ION, RNase III inhibitor | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5M3E
 
 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | Descriptor: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ariza, A, Fontana, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
8COB
 
 | |
6G28
 
 | | Human [protein ADP-ribosylargenine] hydrolase ARH1 in complex with ADP-ribose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, ... | | Authors: | Ariza, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
6G2A
 
 | | Human [protein ADP-ribosylargenine] hydrolase ARH1 in complex with ADP-HPM | | Descriptor: | ACETATE ION, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, CHLORIDE ION, ... | | Authors: | Ariza, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
6G1P
 
 | | Apo form of ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae | | Descriptor: | ACETATE ION, ADP-ribosylhydrolase like 2, CITRIC ACID, ... | | Authors: | Ariza, A. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
6G1Q
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with ADP-ribose | | Descriptor: | ADP-ribosylhydrolase like 2, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ariza, A. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
6HOZ
 
 | |
4ARO
 
 | | Hafnia Alvei phytase in complex with myo-inositol hexakis sulphate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, DI(HYDROXYETHYL)ETHER, HISTIDINE ACID PHOSPHATASE, ... | | Authors: | Moroz, O.V, Blagova, E.B, Ariza, A, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
6N4C
 
 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FJJ
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2WBA
 
 | | Properties of Trypanothione Reductase From T. brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Jones, D, Ariza, A, Chow, W.H.A, Oza, S.L, Fairlamb, A.H. | | Deposit date: | 2009-02-24 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative Structural, Kinetic and Inhibitor Studies of Trypanosoma Brucei Trypanothione Reductase with T. Cruzi.
Mol.Biochem.Parasitol., 169, 2010
|
|
2WWT
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | INTRACELLULAR SUBTILISIN PROTEASE, SODIUM ION | | Authors: | Vevodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2009-10-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of an Intracellular Subtilisin Reveals Novel Structural Features Unique to This Subtilisin Family.
Structure, 18, 2010
|
|
2X8J
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | GLYCEROL, INTRACELLULAR SUBTILISIN PROTEASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Vedodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an intracellular subtilisin reveals novel structural features unique to this subtilisin family.
Structure, 18, 2010
|
|
6SAO
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
