7DIH
 
 | |
7F30
 
 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Y
 
 | | Crystal structure of OxdB E85A mutant (form I) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Z
 
 | | Crystal structure of OxdB E85A mutant (form II) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
6JSA
 
 | |
6JS9
 
 | |
6JSC
 
 | |
6JSD
 
 | |
6JSB
 
 | |
2FMY
 
 | |
7DVU
 
 | | Crystal structure of heme sensor protein PefR in complex with heme and cyanide | | Descriptor: | CYANIDE ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVT
 
 | | Crystal structure of heme sensor protein PefR in complex with heme and carbon monoxide | | Descriptor: | CARBON MONOXIDE, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVR
 
 | | Crystal structure of heme sensor protein PefR from Streptococcus agalactiae in complex with heme | | Descriptor: | COBALT (II) ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVV
 
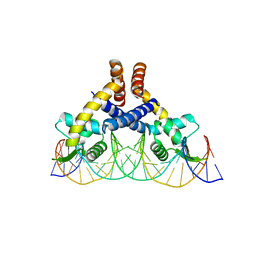 | | Heme sensor protein PefR from Streptococcus agalactiae bound to operator DNA (28-mer) | | Descriptor: | DNA (28-MER), HTH marR-type domain-containing protein | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
1V5H
 
 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
1ZRP
 
 | | SOLUTION-STATE STRUCTURE BY NMR OF ZINC-SUBSTITUTED RUBREDOXIN FROM THE MARINE HYPERTHERMOPHILIC ARCHAEBACTERIUM PYROCOCCUS FURIOSUS | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Blake, P.R, Park, J.B, Zhou, Z.H, Hare, D.R, Adams, M.W.W, Summers, M.F. | | Deposit date: | 1992-07-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus.
Protein Sci., 1, 1992
|
|
