4Z2Q
 
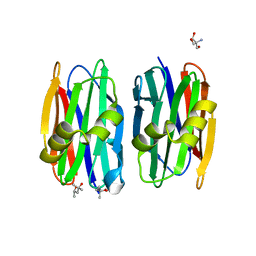 | | The crystal structure of Sclerotium Rolfsii lectin variant 1 (SSR1) in complex with N-acetyl-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
8UD0
 
 | |
8UCZ
 
 | |
8UCY
 
 | |
5HKW
 
 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
2M1B
 
 | |
4KAV
 
 | |
5QTZ
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[1-(2,2-DIFLUOROETHYL)-4-(6-METHYLPYRIDIN-2-YL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-A]PYRIDINE | | Descriptor: | 6-[1-(2,2-difluoroethyl)-4-(6-methylpyridin-2-yl)-1H-imidazol-5-yl]imidazo[1,2-a]pyridine, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5QU0
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[4-(3-CHLORO-4-FLUOROPHENYL)-1-(2-HYDROXYETHYL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-B]PYRIDAZINE-3-CARBONITRILE | | Descriptor: | 6-[4-(3-chloro-4-fluorophenyl)-1-(2-hydroxyethyl)-1H-imidazol-5-yl]imidazo[1,2-b]pyridazine-3-carbonitrile, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
4YLD
 
 | | The crystal structure of Sclerotium Rolfsii lectin variant 1 (SSR1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Sclerotium Rolfsii lectin variant 1 (SSR1) | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
4Z2S
 
 | | The crystal structure of Sclerotium Rolfsii lectin variant 2 (SSR2) in complex with N-acetyl-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
4Z2F
 
 | | Sclerotium Rolfsii lectin variant (SSR2) with fine carbohydrate specificity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, lectin variant 2 | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
