2JWU
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Gb88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-25 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KDM
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | DESIGNED PROTEIN | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From the Cover: A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KDL
 
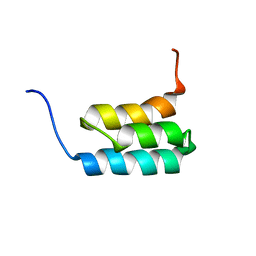 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | designed protein | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2LHE
 
 | | Gb98-T25I,L20A | | Descriptor: | Gb98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHG
 
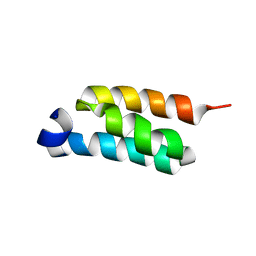 | | GB98-T25I solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
8SZX
 
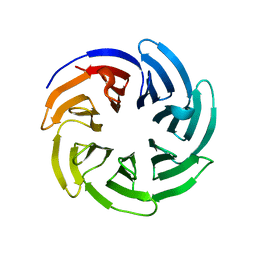 | |
8ENS
 
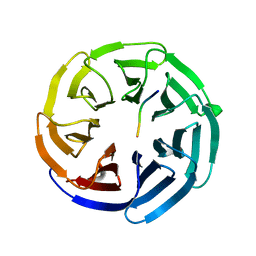 | |
8ENW
 
 | |
8ENY
 
 | |
8ENZ
 
 | |
8ENX
 
 | |
8EO0
 
 | |
