2ZCI
 
 | |
2VC6
 
 | | Structure of MosA from S. meliloti with pyruvate bound | | Descriptor: | DIHYDRODIPICOLINATE SYNTHASE | | Authors: | Phenix, C.P, Nienaber, K.H, Tam, P.H, Delbaere, L.T.J, Palmer, D.R.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, functional and calorimetric investigation of MosA, a dihydrodipicolinate synthase from Sinorhizobium meliloti l5-30, does not support involvement in rhizopine biosynthesis.
Chembiochem, 9, 2008
|
|
1LIN
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:4 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Vandonselaar, M, Hickie, R.A, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trifluoperazine-induced conformational change in Ca(2+)-calmodulin.
Nat.Struct.Biol., 1, 1994
|
|
1CM3
 
 | | HIS15ASP HPR FROM E. COLI | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Waygood, E.B, Delbaere, L.T.J. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
1CM2
 
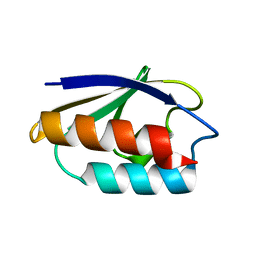 | | STRUCTURE OF HIS15ASP HPR AFTER HYDROLYSIS OF RINGED SPECIES. | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Delbaere, L.T.J, Waygood, E.B. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
2ULL
 
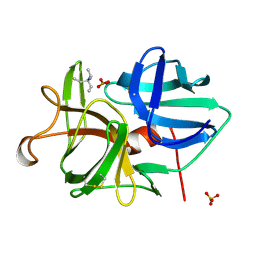 | |
1K3D
 
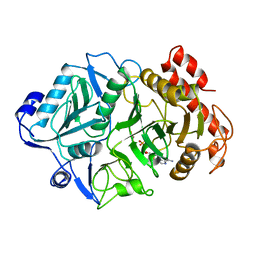 | | Phosphoenolpyruvate carboxykinase in complex with ADP and AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1K3C
 
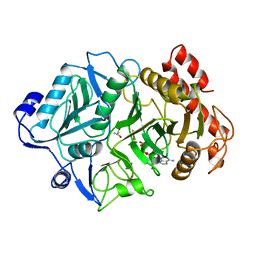 | | Phosphoenolpyruvate carboxykinase in complex with ADP, AlF3 and Pyruvate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1TAL
 
 | | ALPHA-LYTIC PROTEASE AT 120 K (SINGLE STRUCTURE MODEL) | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Rader, S.D, Agard, D.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational substates in enzyme mechanism: the 120 K structure of alpha-lytic protease at 1.5 A resolution.
Protein Sci., 6, 1997
|
|
1FU0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PHOSPHO-SERINE 46 HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Audette, G.F, Engelmann, R, Hengstenberg, W, Deutscher, J, Hayakawa, K, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A resolution structure of phospho-serine 46 HPr from Enterococcus faecalis.
J.Mol.Biol., 303, 2000
|
|
1FX5
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ULEX EUROPAEUS LECTIN I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ANTI-H(O) LECTIN I, CALCIUM ION, ... | | Authors: | Audette, G.F, Vandonselaar, M, Delbaere, L.T.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of the O(H) blood-group-specific lectin I from Ulex europaeus.
J.Mol.Biol., 304, 2000
|
|
2GL0
 
 | | Structure of PAE2307 in complex with adenosine | | Descriptor: | ADENOSINE, PHOSPHATE ION, conserved hypothetical protein | | Authors: | Lott, J.S, Paget, B, Johnston, J.M, Baker, E.N. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Ancient Conserved Domain Establishes a Structural Basis for Stable Histidine Phosphorylation and Identifies a New Family of Adenosine-specific Kinases.
J.Biol.Chem., 281, 2006
|
|
1BOQ
 
 | | PRO REGION C-TERMINUS: PROTEASE ACTIVE SITE INTERACTIONS ARE CRITICAL IN CATALYZING THE FOLDING OF ALPHA-LYTIC PROTEASE | | Descriptor: | PROTEIN (ALPHA-LYTIC PROTEASE), SULFATE ION | | Authors: | Peters, R.J, Shiau, A.K, Sohl, J.L, Anderson, D.E, Tang, G, Silen, J.L, Agard, D.A. | | Deposit date: | 1998-08-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pro region C-terminus:protease active site interactions are critical in catalyzing the folding of alpha-lytic protease.
Biochemistry, 37, 1998
|
|
1OEN
 
 | | PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Descriptor: | ACETATE ION, PHOSPHOENOLPYRUVATE CARBOXYKINASE | | Authors: | Matte, A, Goldie, H, Sweet, R.M, Delbaere, L.T.J. | | Deposit date: | 1995-09-08 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold.
J.Mol.Biol., 256, 1996
|
|
1PTF
 
 | |
1POH
 
 | |
1OPD
 
 | |
1QY6
 
 | | Structue of V8 Protease from Staphylococcus aureus | | Descriptor: | POTASSIUM ION, serine protease | | Authors: | Prasad, L, Leduc, Y, Hayakawa, K, Delbaere, L.T.J. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | Deposit date: | 1999-04-13 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
