8B3X
 
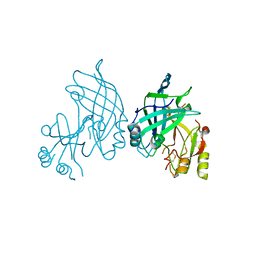 | | High resolution crystal structure of dimeric SUDV VP40 | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Norris, M, Saphire, E.O, Becker, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
6GEL
 
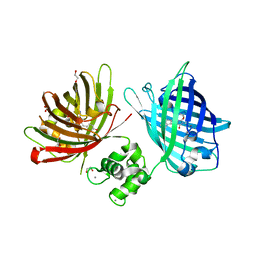 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
5ABS
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL COILED-COIL DOMAIN OF CIN85 IN SPACE GROUP P321 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1, ZINC ION | | Authors: | Wong, L, Habeck, M, Griesinger, C, Becker, S. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Adaptor Protein Cin85 Assembles Intracellular Signaling Clusters for B Cell Activation.
Sci.Signal., 9, 2016
|
|
1G8X
 
 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
5NWM
 
 | |
4C4X
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Z
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Y
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A4 | | Descriptor: | 1-(3-chlorophenyl)-3-(2-methoxyethyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6EHM
 
 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
3UFE
 
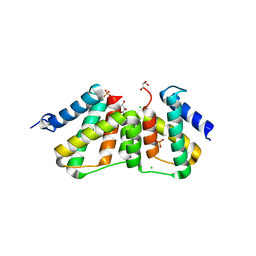 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
6EHL
 
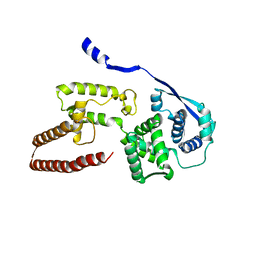 | | Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies | | Descriptor: | Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
6I42
 
 | |
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | Descriptor: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | Authors: | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
3EE6
 
 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
2W0N
 
 | | Plasticity of PAS domain and potential role for signal transduction in the histidine-kinase DcuS | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Etzkorn, M, Kneuper, H, Duennwald, P, Vijayan, V, Kraemer, J, Griesinger, C, Becker, S, Unden, G, Baldus, M. | | Deposit date: | 2008-08-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Plasticity of the Pas Domain and a Potential Role for Signal Transduction in the Histidine Kinase Dcus.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8OWJ
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWE
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L1 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWD
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XIU
 
 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
