2YS4
 
 | | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human | | Descriptor: | Hydrocephalus-inducing protein homolog | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human
To be Published
|
|
2YX7
 
 | | Crystals structure of T132A mutant of St1022 from sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2YYB
 
 | | Crystal structure of TTHA1606 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical protein TTHA1606 | | Authors: | Tomoike, F, Nakagwa, N, Ebihara, A, Yokoyama, S, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TTHA1606 from Thermus thermophilus HB8.
Proteins, 76, 2009
|
|
2YQR
 
 | | Solution structure of the KH domain in KIAA0907 protein | | Descriptor: | KIAA0907 protein | | Authors: | Kadirvel, S, He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KH domain in KIAA0907 protein
To be Published
|
|
2YRL
 
 | |
2YVA
 
 | | Crystal structure of Escherichia coli DiaA | | Descriptor: | DnaA initiator-associating protein diaA | | Authors: | Keyamura, K, Fujikawa, N, Ishida, T, Ozaki, S, Suetsugu, M, Kagawa, W, Yokoyama, S, Kurumizaka, H, Katayama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes
Genes Dev., 21, 2007
|
|
2Z0J
 
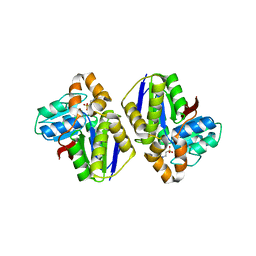 | | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Putative uncharacterized protein TTHA1438 | | Authors: | Nakagawa, N, Kukimoto-Niino, M, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermus thermophilus HB8
To be Published
|
|
2YSK
 
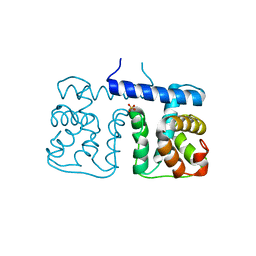 | |
2YWI
 
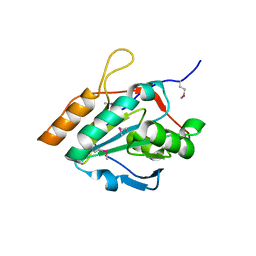 | |
2YT8
 
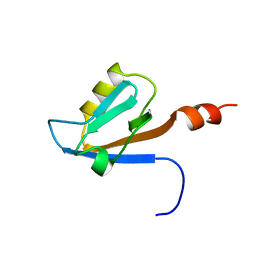 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma)
To be Published
|
|
2YXH
 
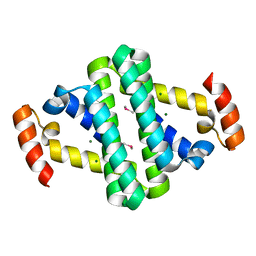 | |
2YWA
 
 | |
2YX5
 
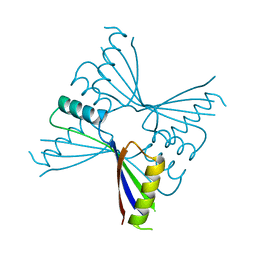 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2Z13
 
 | | Crystal structure of the N-terminal DUF1126 in human EF-hand domain | | Descriptor: | EF-hand domain-containing family member C2 | | Authors: | Saito, K, Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the N-terminal DUF1126 in human EF-hand domain
To be Published
|
|
3AAL
 
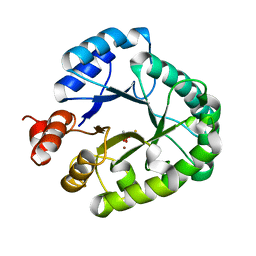 | | Crystal Structure of endonuclease IV from Geobacillus kaustophilus | | Descriptor: | CACODYLATE ION, FE (III) ION, Probable endonuclease 4, ... | | Authors: | Asano, R, Ishikawa, H, Nakane, S, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-11-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An additional C-terminal loop in endonuclease IV, an apurinic/apyrimidinic endonuclease, controls binding affinity to DNA
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2YX6
 
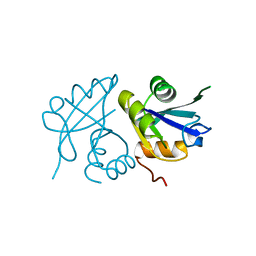 | | Crystal structure of PH0822 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hypothetical protein PH0822 | | Authors: | Hosaka, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0822
To be Published
|
|
2YZS
 
 | |
2Z08
 
 | |
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ACC
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with GMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2YVT
 
 | |
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ACB
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2YZI
 
 | |
2YZJ
 
 | | Crystal structure of dCTP deaminase from Sulfolobus tokodaii | | Descriptor: | 167aa long hypothetical dUTPase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of dCTP deaminase from Sulfolobus tokodaii
To be Published
|
|
