5XPV
 
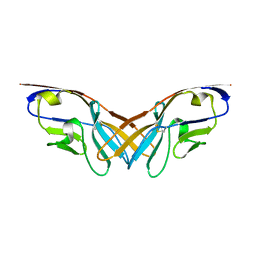 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5ZKX
 
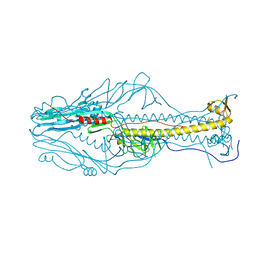 | |
5ZL2
 
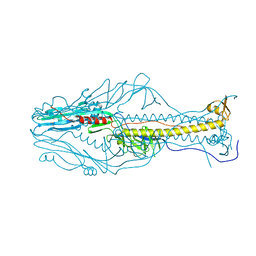 | |
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F03
 
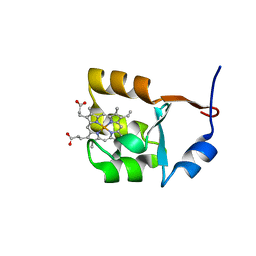 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
3L95
 
 | |
5KWA
 
 | |
8JZV
 
 | | RPA70N-ETAA1 fusion | | Descriptor: | Ewing's tumor-associated antigen 1, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8JZY
 
 | | RPA70N-RAD9 fusion | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8K00
 
 | | RPA70N-MRE11 fusion | | Descriptor: | Double-strand break repair protein MRE11, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
5BOE
 
 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UBD
 
 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
5KZF
 
 | |
7W8O
 
 | |
7W8K
 
 | |
7W8R
 
 | |
7W96
 
 | |
7WE3
 
 | |
7WEI
 
 | |
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7W8Z
 
 | |
7W8T
 
 | |
2R0L
 
 | | Short Form HGFA with Inhibitory Fab75 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
