3NPQ
 
 | |
2LHR
 
 | |
4XLY
 
 | | The complex structure of KS-D75C with substrate CPP | | Descriptor: | (2E)-3-methyl-5-[(1R,4aR,8aR)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
3OTP
 
 | |
4XWF
 
 | |
4XLX
 
 | | Crystal structure of BjKS from Bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XW7
 
 | |
4XP7
 
 | | Crystal structure of Human tRNA dihydrouridine synthase 2 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Whelan, F, Jenkins, H.T, Griffiths, S, Byrne, R.T, Dodson, E.J, Antson, A.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From bacterial to human dihydrouridine synthase: automated structure determination.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2JMF
 
 | | Solution structure of the Su(dx) WW4- Notch PY peptide complex | | Descriptor: | E3 ubiquitin-protein ligase suppressor of deltex, Neurogenic locus Notch protein | | Authors: | Avis, J.M, Blankley, R.T, Jennings, M.D, Golovanov, A.P. | | Deposit date: | 2006-11-05 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and Autoregulation of Notch Binding by Tandem WW Domains in Suppressor of Deltex
J.Biol.Chem., 282, 2007
|
|
4YJM
 
 | |
4YCP
 
 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNATrp | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NPN
 
 | |
4WW2
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8, Beta Chain-TRBV7-8, Antigen-presenting glycoprotein CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, S, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4YKA
 
 | | The structure of Agrobacterium tumefaciens ClpS2 in complex with L-tyrosinamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, L-TYROSINAMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
4WNQ
 
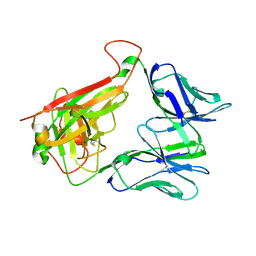 | | THE MOLECULAR BASES OF DELTA/ALPHA-BETA T-CELL MEDIATED ANTIGEN RECOGNITION | | Descriptor: | TCR Variable Beta 2 (TRBV20) chain and TCR constant Beta chain, TCR Variable Delta 1 chain and TCR constant Alpha chain | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4WO4
 
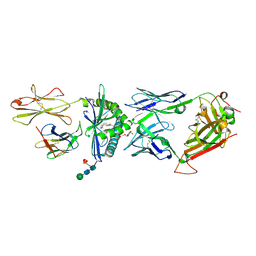 | | The molecular bases of Delta/Alpha beta T cell-mediated antigen recognition. | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
3O2H
 
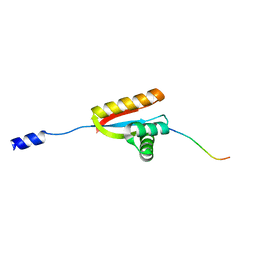 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
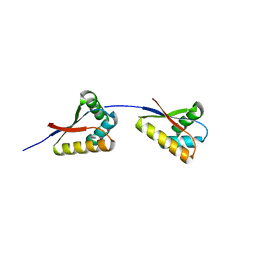 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
4YCO
 
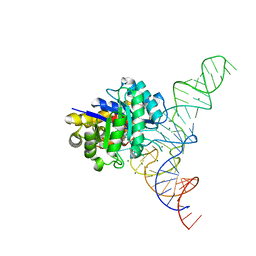 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNAPhe | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LA0
 
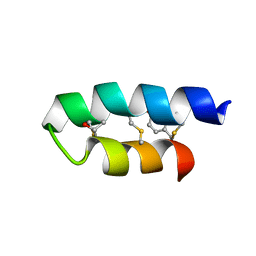 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
2M60
 
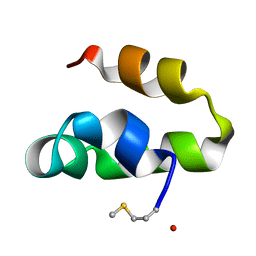 | | Enterocin 7B | | Descriptor: | Enterocin JSB | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
4YJX
 
 | | The structure of Agrobacterium tumefaciens ClpS2 bound to L-phenylalaninamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, PHENYLALANINE AMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
