9C97
 
 | |
9CMR
 
 | | Saccharomyces cerevisiae Tom1 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase TOM1, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P. | | Deposit date: | 2024-07-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tom1p ubiquitin ligase structure, interaction with Spt6p, and function in maintaining normal transcript levels and the stability of chromatin in promoters.
To Be Published
|
|
9E3B
 
 | | Cryo-EM structure of PRMT5/WDR77 in complex with 6S complex | | Descriptor: | Methylosome protein WDR77, Methylosome subunit pICln, Protein arginine N-methyltransferase 5, ... | | Authors: | Jin, C.Y, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2024-10-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Substrate adaptors are flexible tethering modules that enhance substrate methylation by the arginine methyltransferase PRMT5.
J.Biol.Chem., 301, 2025
|
|
9DGZ
 
 | |
9DH0
 
 | | The Cryo-EM structure of recombinantly expressed hUGDH in complex with UDP-4-keto-xylose | | Descriptor: | (2R,3R,4R)-3,4-dihydroxy-5-oxooxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), UDP-glucose 6-dehydrogenase | | Authors: | Kadirvelraj, R, Walsh Jr, R.M, Wood, Z.W. | | Deposit date: | 2024-09-03 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM Structure of Recombinantly Expressed hUGDH Unveils a Hidden, Alternative Allosteric Inhibitor.
Biochemistry, 64, 2025
|
|
9EGW
 
 | |
9EGV
 
 | | HOIL-1 RING2 domain bound to ubiquitin | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The RBR E3 ubiquitin ligase HOIL-1 can ubiquitinate diverse non-protein substrates in vitro.
Life Sci Alliance, 8, 2025
|
|
9EC2
 
 | | Crystal structure of SAMHD1 dimer bound to an inhibitor obtained from high-throughput chemical tethering to the guanine antiviral acyclovir | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, N-[5-({2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl}amino)-5-oxopentyl]-4,7-dibromo-3-hydroxynaphthalene-2-carboxamide | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Lopez-Rovira, L.M, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2024-11-13 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inhibitors of SAMHD1 Obtained from Chemical Tethering to the Guanine Antiviral Acyclovir.
Biochemistry, 64, 2025
|
|
9DYB
 
 | |
9DYA
 
 | | CHIP-TPR in complex with the Hsp70 tail | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock 70 kDa protein 1A, SULFATE ION | | Authors: | Zhang, H, Nix, J.C, Page, R.C. | | Deposit date: | 2024-10-13 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Phosphorylation-State Modulated Binding of HSP70: Structural Insights and Compensatory Protein Engineering.
Biorxiv, 2025
|
|
9DJT
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue WIZ-5 | | Descriptor: | (3S)-3-(5-{[(4R)-6-ethyl-6-azaspiro[2.5]octan-4-yl]oxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, DNA damage-binding protein 1, Protein Wiz, ... | | Authors: | Partridge, J.R, Ma, X, Ornelas, E. | | Deposit date: | 2024-09-06 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery and Optimization of First-in-Class Molecular Glue Degraders of the WIZ Transcription Factor for Fetal Hemoglobin Induction to Treat Sickle Cell Disease.
J.Med.Chem., 67, 2024
|
|
9E6S
 
 | |
9E6T
 
 | |
9C12
 
 | | Leukotriene C4-bound Multidrug Resistance-associated Protein 2 (MRP2) | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL, ... | | Authors: | Koide, E, Chen, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for the transport and regulation mechanism of the multidrug resistance-associated protein 2.
Nat Commun, 16, 2025
|
|
9B9P
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, and JK-5-114 | | Descriptor: | 6-hydroxy-2-phenyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Lin, X, Guddat, L.W. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | A Ketol-Acid Reductoisomerase Inhibitor That Has Antituberculosis and Herbicidal Activity.
Chemistry, 31, 2025
|
|
9D7I
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 KN Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7H
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 KN Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7G
 
 | | BG505 DS-SOSIP.664 apo structure from the CH103 KN cryo-EM dataset | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface protein gp120, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7O
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7P
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9C6M
 
 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase, K73M mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA ligase1 | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9CE6
 
 | |
9C6L
 
 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, RNA ligase1, ... | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | 1.94 angstrom structure of synthetic alpha-synuclein fibrils seeding MSA neuropathology
Biorxiv, 2024
|
|
9DIK
 
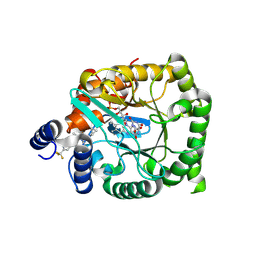 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
