7WT3
 
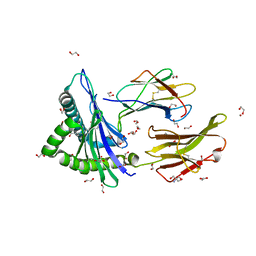 | | Crystal structure of HLA-A*2402 complexed with 4-mer lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-mer lipopeptide, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.887655 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
1IX1
 
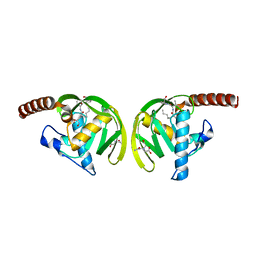 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
6IWG
 
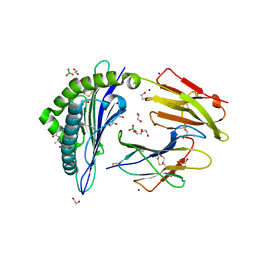 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
6IWH
 
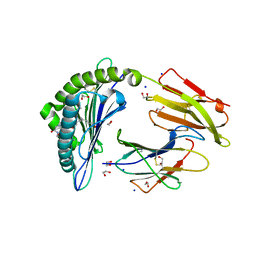 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6JHX
 
 | |
6LB2
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with mono-acyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69380951 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
2ZBI
 
 | |
2ZZR
 
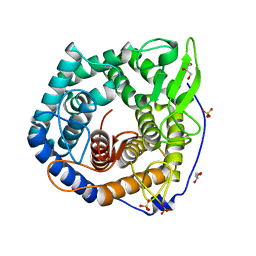 | |
2ZK9
 
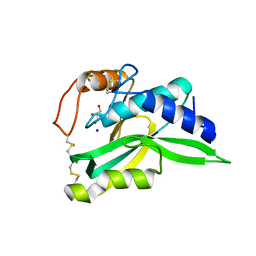 | | Crystal Structure of Protein-glutaminase | | Descriptor: | GLYCEROL, Protein-glutaminase, SODIUM ION | | Authors: | Hashizume, R. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-17 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of protein glutaminase and its pro forms converted into enzyme-substrate complex
J.Biol.Chem., 286, 2011
|
|
1LLD
 
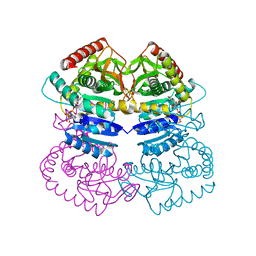 | |
3SWE
 
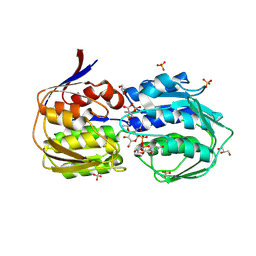 | | Haemophilus influenzae MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys117 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
1XGF
 
 | |
3WXS
 
 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
