9DIK
 
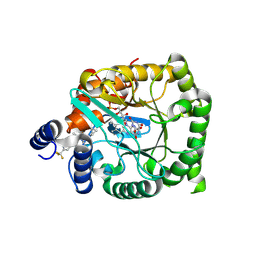 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9CS8
 
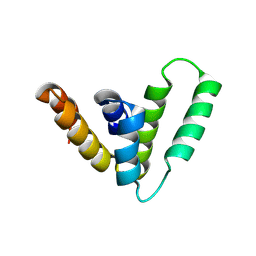 | | Structure of Azospirillum bacterial CARD crystal form 2 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, Hill, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
9CS7
 
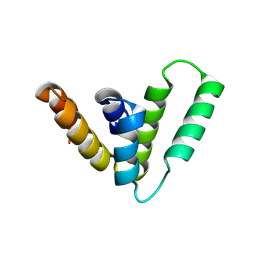 | | Structure of Azospirillum bacterial CARD crystal form 1 | | Descriptor: | Serine protease | | Authors: | Wein, T, Millman, A, Lange, K, Yirmiya, E, Hadary, R, Garb, J, Melamed, S, Steinruecke, F, HIll, A.B, Kranzusch, P.J, Sorek, R. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CARD domains mediate anti-phage defence in bacterial gasdermin systems.
Nature, 639, 2025
|
|
9DJT
 
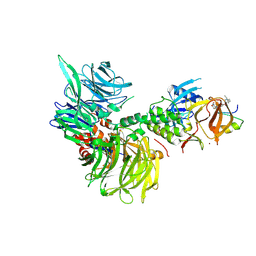 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue WIZ-5 | | Descriptor: | (3S)-3-(5-{[(4R)-6-ethyl-6-azaspiro[2.5]octan-4-yl]oxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, DNA damage-binding protein 1, Protein Wiz, ... | | Authors: | Partridge, J.R, Ma, X, Ornelas, E. | | Deposit date: | 2024-09-06 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery and Optimization of First-in-Class Molecular Glue Degraders of the WIZ Transcription Factor for Fetal Hemoglobin Induction to Treat Sickle Cell Disease.
J.Med.Chem., 67, 2024
|
|
9DGZ
 
 | |
9DH0
 
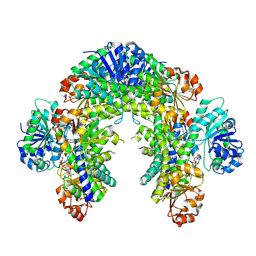 | | The Cryo-EM structure of recombinantly expressed hUGDH in complex with UDP-4-keto-xylose | | Descriptor: | (2R,3R,4R)-3,4-dihydroxy-5-oxooxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), UDP-glucose 6-dehydrogenase | | Authors: | Kadirvelraj, R, Walsh Jr, R.M, Wood, Z.W. | | Deposit date: | 2024-09-03 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM Structure of Recombinantly Expressed hUGDH Unveils a Hidden, Alternative Allosteric Inhibitor.
Biochemistry, 64, 2025
|
|
9CE6
 
 | |
9CYB
 
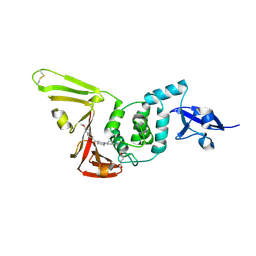 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P1 | | Descriptor: | Papain-like protease, SUCCINIC ACID, [(3R)-1-cyclopentylpiperidin-3-yl](6-methoxynaphthalen-2-yl)methanone | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYK
 
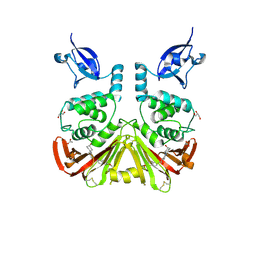 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P24 | | Descriptor: | ACETIC ACID, GLYCEROL, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYC
 
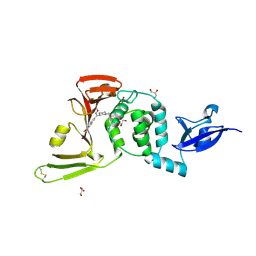 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P2 | | Descriptor: | (E)-1-[(3R)-1-cyclopentylpiperidin-3-yl]-N-methoxy-1-(6-methoxynaphthalen-2-yl)methanimine, ACETIC ACID, GLYCEROL, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchel, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYD
 
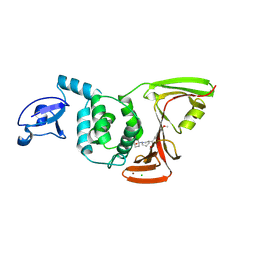 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P4 | | Descriptor: | (1S,4s)-4-{(3R)-3-[(E)-(methoxyimino)(6-methoxynaphthalen-2-yl)methyl]piperidin-1-yl}cyclohexan-1-ol, CHLORIDE ION, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CMR
 
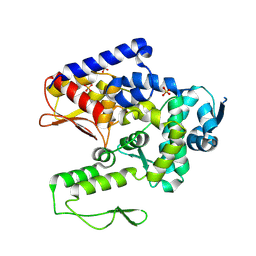 | | Saccharomyces cerevisiae Tom1 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase TOM1, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P. | | Deposit date: | 2024-07-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tom1p ubiquitin ligase structure, interaction with Spt6p, and function in maintaining normal transcript levels and the stability of chromatin in promoters.
To Be Published
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
9EM1
 
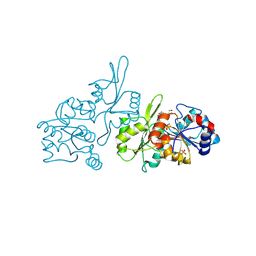 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
9F18
 
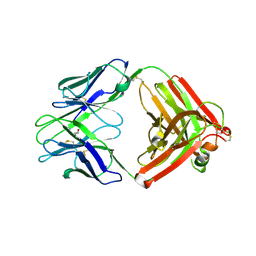 | |
9EQF
 
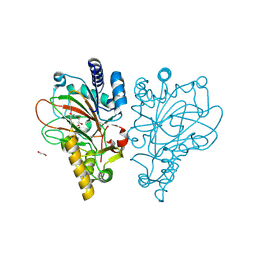 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
9EGV
 
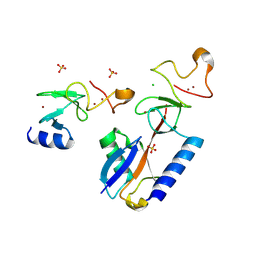 | | HOIL-1 RING2 domain bound to ubiquitin | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The RBR E3 ubiquitin ligase HOIL-1 can ubiquitinate diverse non-protein substrates in vitro.
Life Sci Alliance, 8, 2025
|
|
9ETL
 
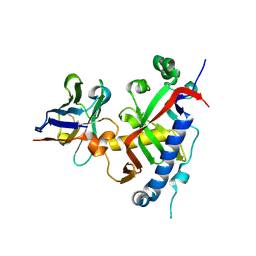 | | Mouse CNPase catalytic domain with nanobody 8D | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, Chains: D,C | | Authors: | Schroder, M, Markusson, S, Raasakka, A, Opazo, F, Kursula, P. | | Deposit date: | 2024-03-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mouse CNPase catalytic domain with nanobody 8D
To Be Published
|
|
9EGW
 
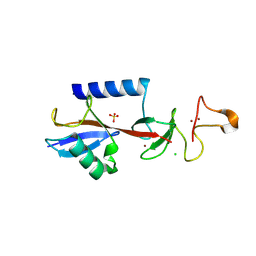 | |
9EOI
 
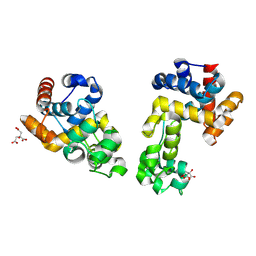 | |
9EOL
 
 | |
9E3B
 
 | | Cryo-EM structure of PRMT5/WDR77 in complex with 6S complex | | Descriptor: | Methylosome protein WDR77, Methylosome subunit pICln, Protein arginine N-methyltransferase 5, ... | | Authors: | Jin, C.Y, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2024-10-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Substrate adaptors are flexible tethering modules that enhance substrate methylation by the arginine methyltransferase PRMT5.
J.Biol.Chem., 301, 2025
|
|
9EWF
 
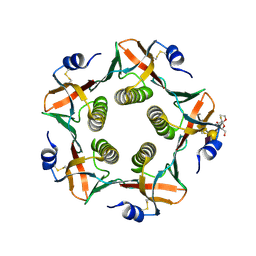 | | Cholera toxin B subunit in complex with fluorinated GM1 | | Descriptor: | (2R,3S,4S,5R,6R)-6-dodecoxy-5-fluoranyl-2-(hydroxymethyl)oxane-3,4-diol, 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, Cholera enterotoxin subunit B | | Authors: | Fan, J, Koehnke, J. | | Deposit date: | 2024-04-03 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Origin of Affinity in the GM1-Cholera Toxin Complex through Site-Selective Editing with Fluorine.
Acs Cent.Sci., 10, 2024
|
|
9EU4
 
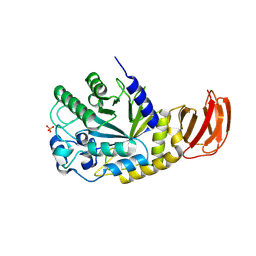 | | GH29A alpha-L-fucosidase | | Descriptor: | Exported alpha-L-fucosidase protein, SULFATE ION, beta-L-fucopyranose | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
9EU3
 
 | | GH29A alpha-L-fucosidase | | Descriptor: | Alpha-L-fucosidase, CHLORIDE ION, ZINC ION, ... | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
