7OX4
 
 | | Mouse interleukin-9 in complex with Fab 35D8. | | Descriptor: | ACETATE ION, Heavy chain (Fab 35D8), Interleukin-9, ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
8P8K
 
 | | Acyl-ACP thioesterase from Lemna paucicostata in complex with a thiazolopyridine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2,6-bis(fluoranyl)phenyl]-6-chloranyl-[1,3]thiazolo[4,5-b]pyridine, Acyl-ACP thioesterase | | Authors: | Freigang, J. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Study in Scaffold Hopping: Discovery and Optimization of Thiazolopyridines as Potent Herbicides That Inhibit Acyl-ACP Thioesterase.
J.Agric.Food Chem., 71, 2023
|
|
7EHP
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, chitin oligosaccahride binding protein NagB1 | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHO
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | Chitin oligosaccharide binding protein NagB2, TETRAETHYLENE GLYCOL | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHU
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin oligosaccharide binding protein NagB2, DI(HYDROXYETHYL)ETHER | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7EHQ
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin oligosaccharide binding protein NagB2 | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
5B1H
 
 | |
5ZA1
 
 | | Ligand complex of RORgt LBD | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-({[4-(ethylsulfonyl)phenyl]acetyl}amino)phenyl]-2-methyl-N-phenylpropanamide, DIMETHYLFORMAMIDE, ... | | Authors: | Yamamoto, S, Yamaguchi, H. | | Deposit date: | 2018-02-06 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of a potent orally bioavailable retinoic acid receptor-related orphan receptor-gamma-t (ROR gamma t) inhibitor, S18-000003.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5B1I
 
 | |
5DSV
 
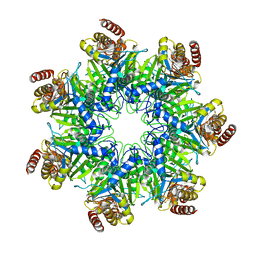 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
3NTY
 
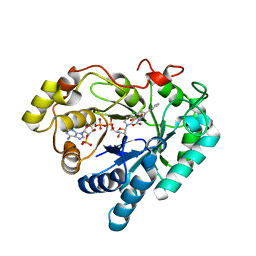 | | Crystal structure of AKR1C1 in complex with NADP and 5-Phenyl,3-chlorosalicylic acid | | Descriptor: | 5-chloro-4-hydroxybiphenyl-3-carboxylic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-06 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the inhibitor selectivity pocket of human 20 alpha-hydroxysteroid dehydrogenase (AKR1C1) with X-ray crystallography and site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1WV3
 
 | |
2Z6F
 
 | |
4HVK
 
 | | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yamanaka, Y, Zeppieri, L, Nicolet, Y, Marinoni, E.N, de Oliveira, J.S, Masafumi, O, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-11-06 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure and functional studies of an unusual L-cysteine desulfurase from Archaeoglobus fulgidus.
Dalton Trans, 42, 2013
|
|
2ZKL
 
 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Yao, M, Tanaka, I, Tsumoto, K. | | Deposit date: | 2008-03-25 | | Release date: | 2009-03-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of capsular polysaccharide assembling protein from Staphylococcus aureus
to be published
|
|
7EKQ
 
 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
3A3G
 
 | | Crystal structure of LumP complexed with 6,7-dimethyl-8-(1'-D-ribityl) lumazine | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Lumazine protein | | Authors: | Sato, Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3VTM
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING INDIUM | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-05-31 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus
Protein Sci., 22, 2013
|
|
3A3B
 
 | | Crystal structure of LumP complexed with flavin mononucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
2ZDI
 
 | | Crystal structure of Prefoldin from Pyrococcus horikoshii OT3 | | Descriptor: | Prefoldin subunit alpha, Prefoldin subunit beta, SULFATE ION | | Authors: | Kida, H, Miki, K. | | Deposit date: | 2007-11-23 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins
J.Mol.Biol., 376, 2008
|
|
3VUA
 
 | | Apo IsdH-NEAT3 in space group P3121 at a resolution of 1.85 A | | Descriptor: | ACETATE ION, GLYCEROL, Iron-regulated surface determinant protein H, ... | | Authors: | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | Deposit date: | 2012-06-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin
To be Published
|
|
3WA6
 
 | |
3A35
 
 | | Crystal structure of LumP complexed with riboflavin | | Descriptor: | Lumazine protein, RIBOFLAVIN | | Authors: | Sato, Y. | | Deposit date: | 2009-06-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1'-D-ribityl) lumazine and its analogues, riboflavin and FMN, at high resolution
J.Bacteriol., 192, 2009
|
|
3APT
 
 | |
