6IQX
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, aerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y3C
 
 | |
5Y3B
 
 | |
3W1F
 
 | | Crystal structure of Human MPS1 catalytic domain in complex with 5-(5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl)-2-methylbenzenesulfonamide | | Descriptor: | 5-[5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl]-2-methylbenzenesulfonamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Tachibana, Y, Itoh, T, Yamamoto, T, Hashizume, H, Hato, Y, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Higaki, M, Ueda, K, Yoshizawa, H, Baba, Y, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Indazole-based potent and cell-active Mps1 kinase inhibitors: rational design from pan-kinase inhibitor anthrapyrazolone (SP600125)
J.Med.Chem., 56, 2013
|
|
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
1INL
 
 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
2ZLY
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZRQ
 
 | | Crystal structure of S324A-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Takeuchi, Y, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-08-28 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Tk-subtilisin folded without propeptide: requirement of propeptide for acceleration of folding
Febs Lett., 582, 2008
|
|
3A20
 
 | |
1UCR
 
 | | Three-dimensional crystal structure of dissimilatory sulfite reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Mizuno, N, Voordouw, G, Miki, K, Sarai, A, Higuchi, Y. | | Deposit date: | 2003-04-18 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Dissimilatory Sulfite Reductase D (DsrD) Protein-Possible Interaction with B- and Z-DNA by Its Winged-Helix Motif
STRUCTURE, 11, 2003
|
|
2D5G
 
 | |
2DOO
 
 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
3A8N
 
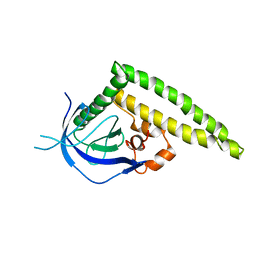 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3AQZ
 
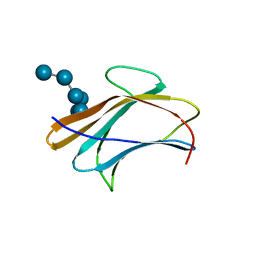 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQY
 
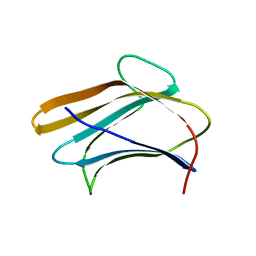 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
6AC1
 
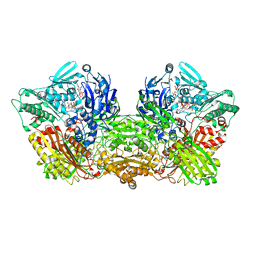 | | Rat Xanthine oxidoreductase, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
5WYA
 
 | | Structure of amino acid racemase, 2.65 A | | Descriptor: | (2S,3S)-3-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pentanoic acid, DIMETHYL SULFOXIDE, Isoleucine 2-epimerase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5X1C
 
 | |
6AC4
 
 | | Rat Xanthine oxidoreductase, D428N variant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Xanthine dehydrogenase/oxidase | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
6AJU
 
 | | Rat Xanthine oxidoreductase | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
